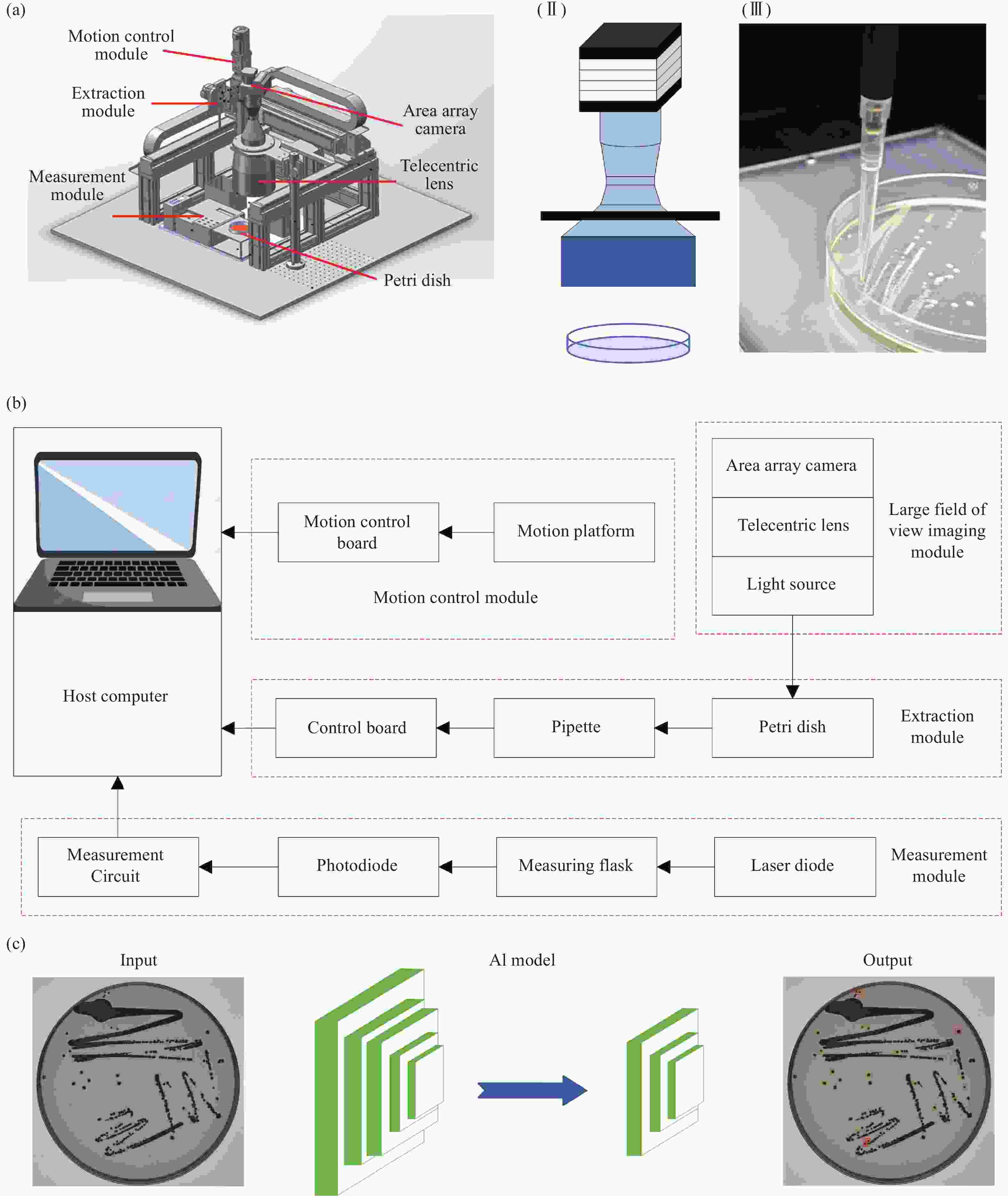
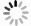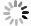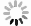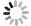Artificial intelligence-enabled high-precision colony extraction and isolation system
doi: 10.37188/CO.EN-2025-0025
-
摘要:
标准菌悬液在微生物诊断中具有重要意义。传统制备方法依赖人工操作,存在重复性差、效率低及生物安全隐患等问题。本研究提出一种融合大视野成像与人工智能技术的高精度自动化菌落提取分选系统,实现菌落智能筛查与定位。首先,开发了大视野成像系统,可采集90 mm培养皿高分辨图像,物理分辨率达13.2 μm,成像速度为13帧/秒;其次,应用人工智能技术实现菌落自动识别与定位,支持筛选直径为1.9~2.3 mm的目标菌落;接着设计三轴运动控制平台,配合路径规划算法实现菌落高效提取,采用电动移液器进行精准菌落采集;同时开发菌悬液浓度测量模块,以650 nm激光二极管为光源,实现0.01麦氏浓度(MCF)的测量精度。最终通过大肠杆菌悬液制备验证系统性能,具体为:经17小时培养后分4次提取大肠杆菌,达到系统设定目标浓度。该工作有望实现微生物样本的快速精准制备,显著缩短检测周期,减轻医务人员工作负担。
Abstract:Standard bacterial suspensions play a crucial role in microbiological diagnosis. Traditional preparation methods, which rely heavily on manual operations, face challenges such as poor reproducibility, low efficiency, and biosafety concerns. In this study, we propose a high-precision automated colony extraction and separation system that combines large-field imaging and artificial intelligence (AI) to facilitate intelligent screening and localization of colonies. Firstly, a large-field imaging system was developed to capture high-resolution images of 90 mm Petri dishes, achieving a physical resolution of 13.2 μm and an imaging speed of 13 frames per second. Subsequently, AI technology was employed for the automatic recognition and localization of colonies, enabling the selection of target colonies with diameters ranging from 1.9 to 2.3 mm. Next, a three-axis motion control platform was designed, accompanied by a path planning algorithm for the efficient extraction of colonies. An electronic pipette was employed for accurate colony collection. Additionally, a bacterial suspension concentration measurement module was developed, incorporating a 650 nm laser diode as the light source, achieving a measurement accuracy of 0.01 McFarland concentration (MCF). Finally, the system’s performance was validated through the preparation of an Esckerichia coli (E. coli) suspension. After 17 hours of cultivation, E. coli was extracted four times, achieving the target concentration set by the system. This work is expected to enable rapid and accurate microbial sample preparation, significantly reducing detection cycles and alleviating the workload of healthcare personnel.
-
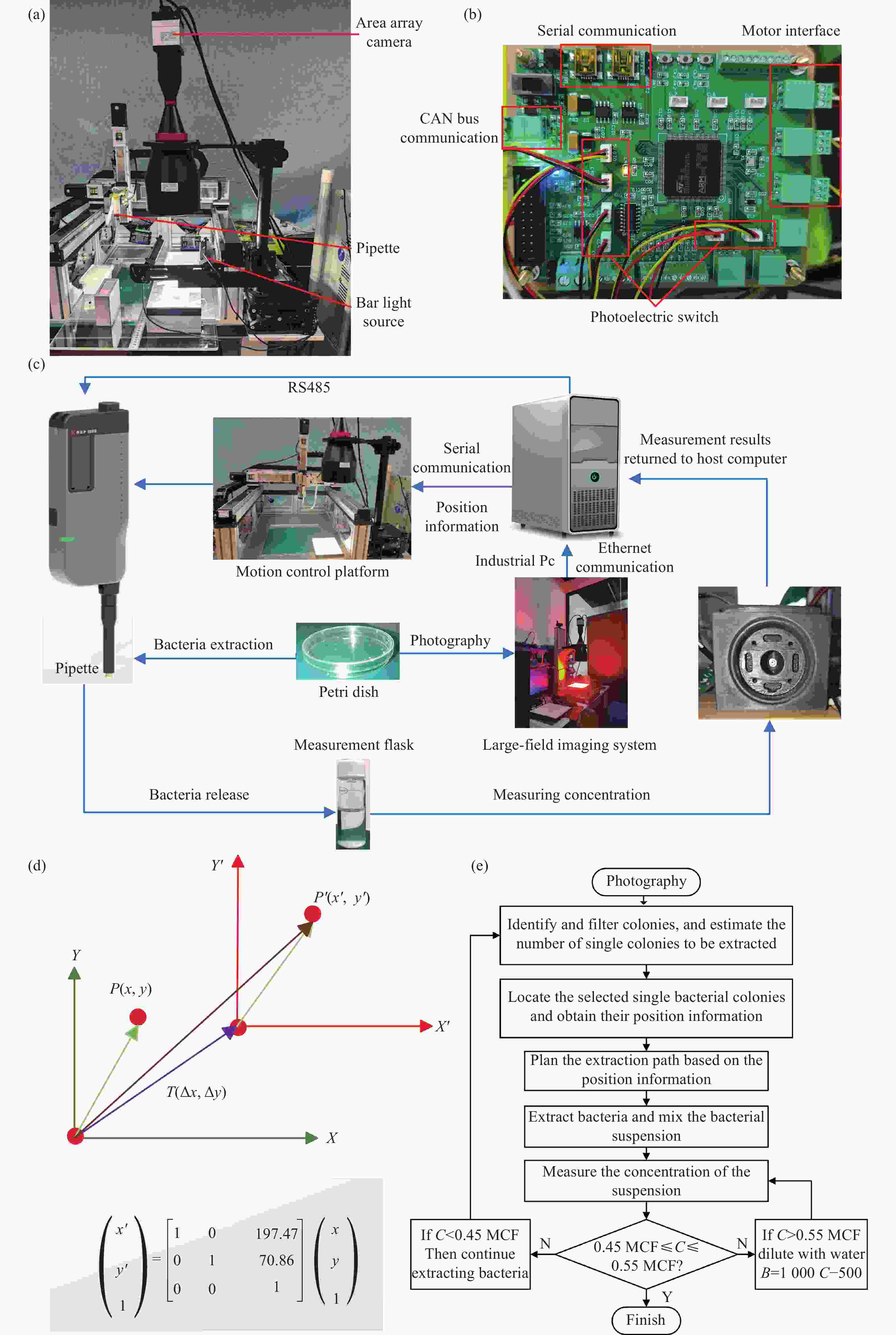
Figure 2. High-precision automated colony extraction system. (a) Core control board. (b) Physical diagram of the optical system for colony extraction. (c) Components of the extraction system. (d) Translation transformation between the world coordinate system and the camera coordinate system. (e) Schematic diagram of dynamic solution adjustment
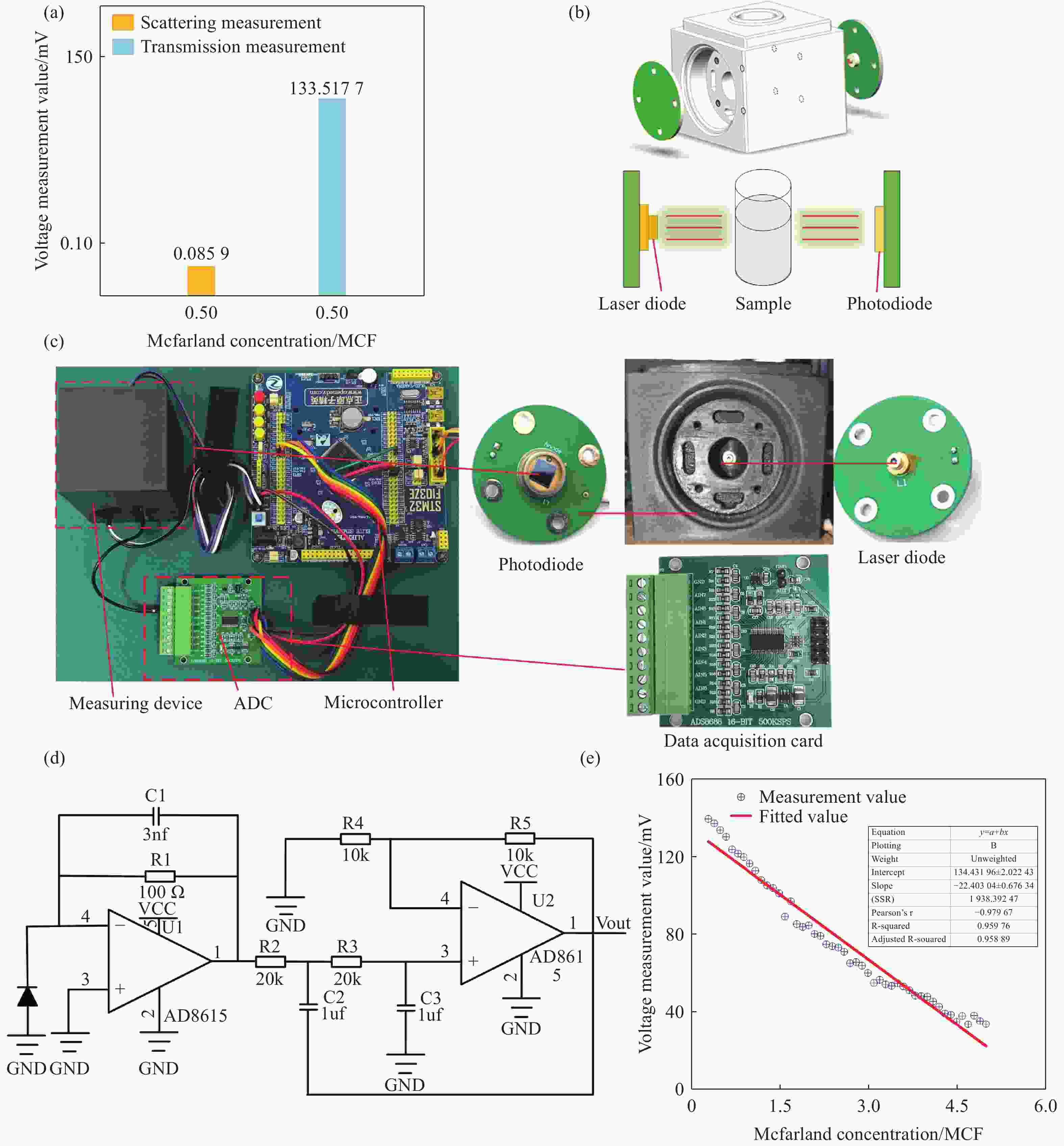
Figure 3. Bacterial suspension concentration measurement system. (a) Comparison of scattering and transmission measurement values at 0.5 McFarland concentration. (b) Schematic diagram and 3D model of the bacterial suspension concentration measurement module. (c) Physical diagram of the single-wavelength measurement system. (d) Circuit diagram of the signal amplification for measurement. (e) Calibration curve of measured voltage versus standard concentration
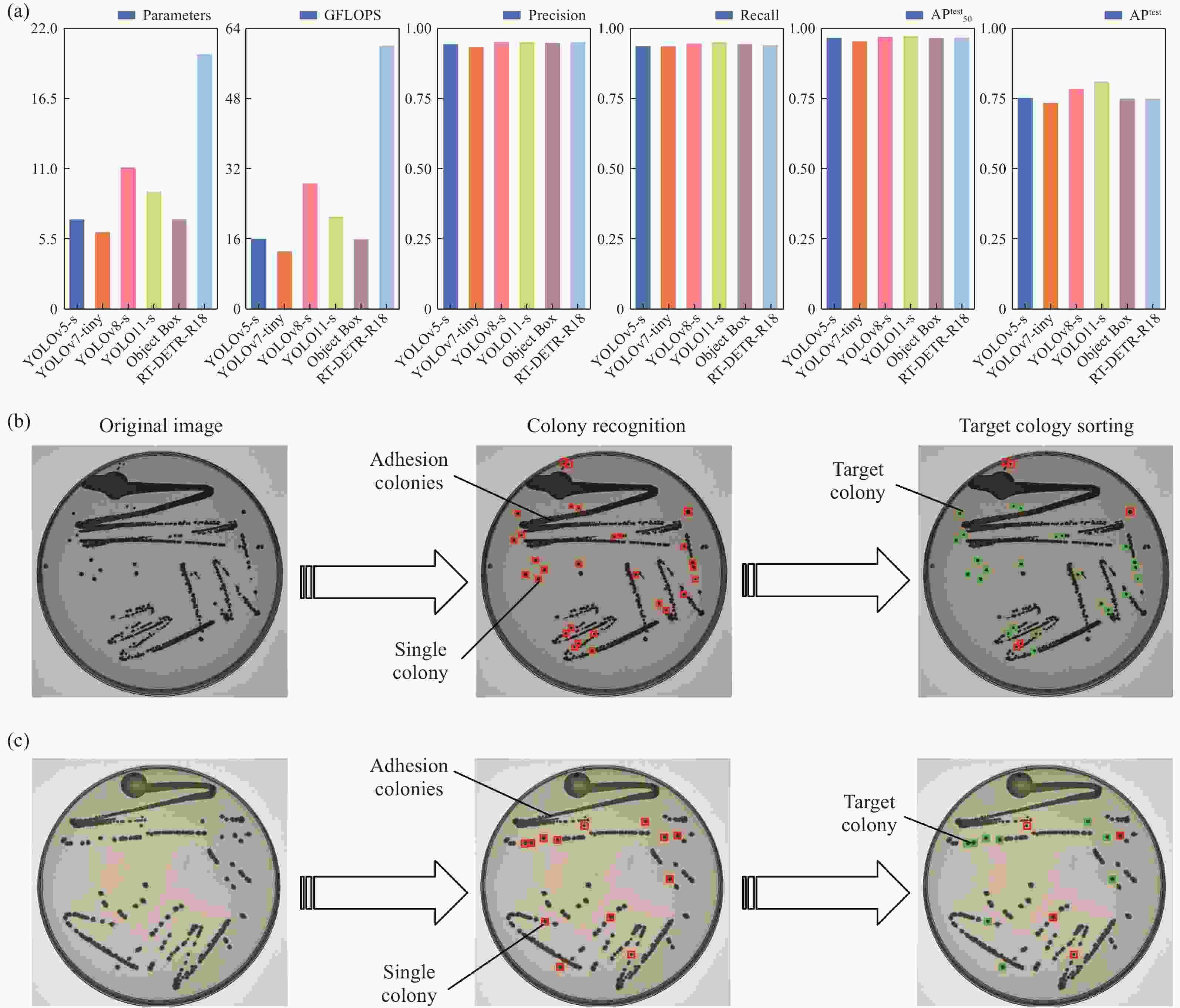
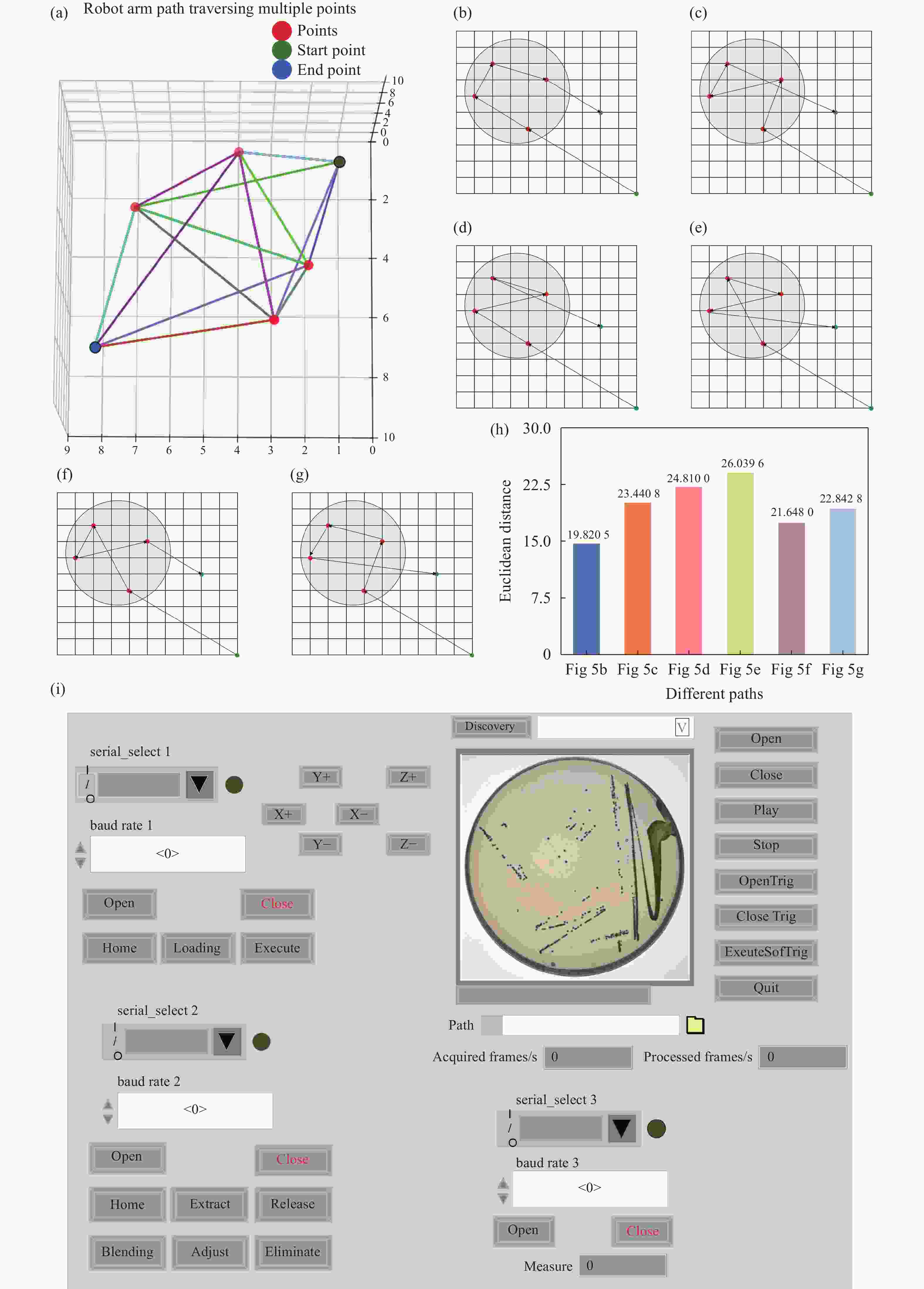
Figure 5. Path planning for single colony extraction. (a) 3D random path diagram. (b) This work. (c) Random path 1. (d) Random path 2. (e) Random path 3. (f) Random path 4. (g) Random path 5. (h) Comparison of Euclidean distances for different path planning strategies. (i) Host computer operation interface
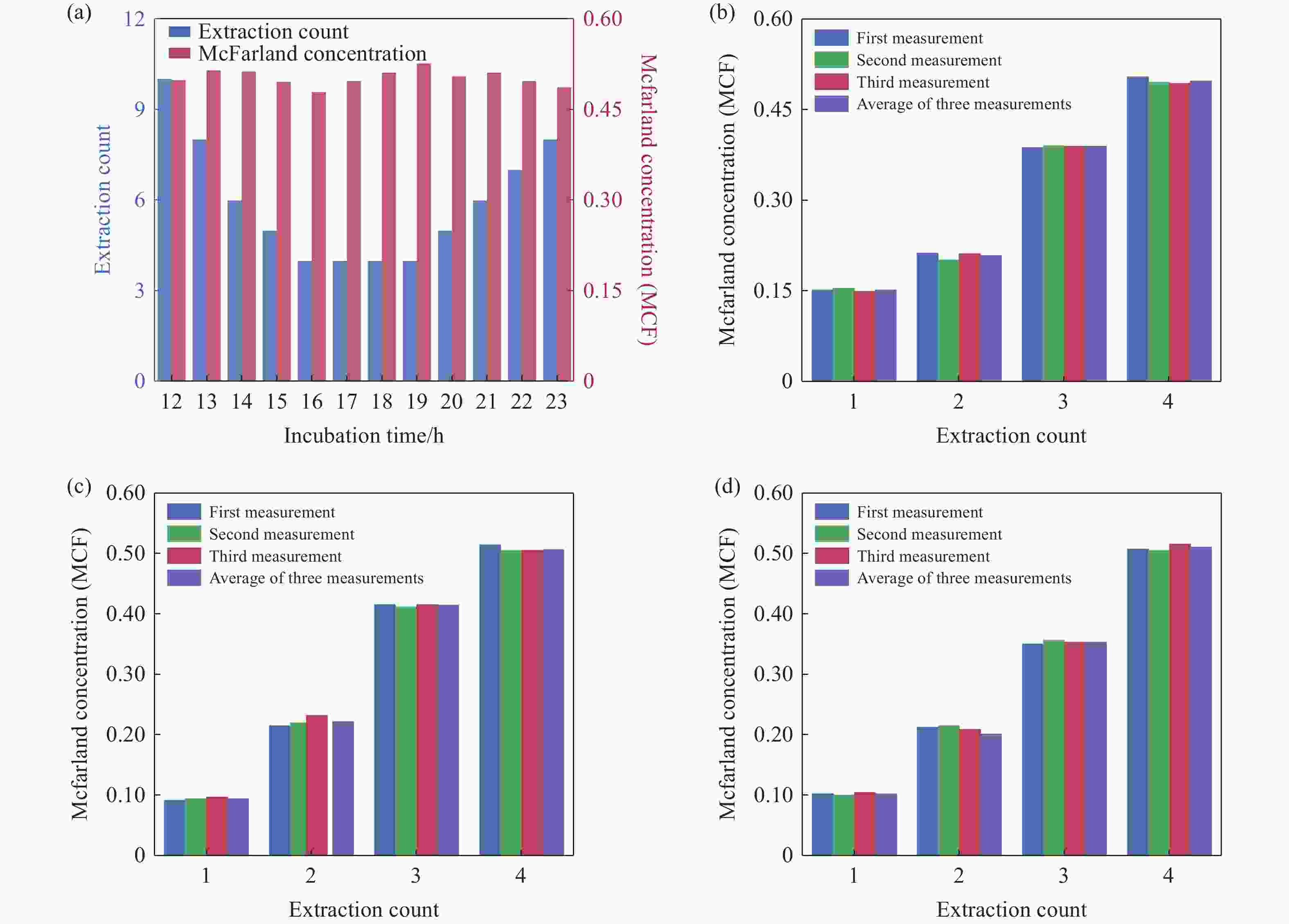
Figure 6. On-demand preparation of E. coli suspension concentration. (a) Relationship between the number of extractions and cultivation time at target concentration. (b-d) Relationship between E. coli concentration and the number of extractions after 17 hours of cultivation: (b) Group 1, (c) Group 2, (d) Group 3
-
[1] OPOTA O, JATON K, GREUB G. Microbial diagnosis of bloodstream infection: towards molecular diagnosis directly from blood[J]. Clinical Microbiology and Infection, 2015, 21(4): 323-331. doi: 10.1016/j.cmi.2015.02.005 [2] DEURENBERG R H, BATHOORN E, CHLEBOWICZ M A, et al. Application of next generation sequencing in clinical microbiology and infection prevention[J]. Journal of Biotechnology, 2017, 243: 16-24. doi: 10.1016/j.jbiotec.2016.12.022 [3] SHELKE Y P, BADGE A K, BANKAR N J. Applications of artificial intelligence in microbial diagnosis[J]. Cureus, 2023, 15(11): e49366. [4] DAVENPORT M, MACH K E, SHORTLIFFE L M D, et al. New and developing diagnostic technologies for urinary tract infections[J]. Nature Reviews Urology, 2017, 14(5): 296-310. doi: 10.1038/nrurol.2017.20 [5] MIAO Q, MA Y Y, WANG Q Q, et al. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice[J]. Clinical Infectious Diseases, 2018, 67(suppl_2): S231-S240. doi: 10.1093/cid/ciy693 [6] GAJIC I, KABIC J, KEKIC D, et al. Antimicrobial susceptibility testing: a comprehensive review of currently used methods[J]. Antibiotics, 2022, 11(4): 427. doi: 10.3390/antibiotics11040427 [7] HU P T, GAO R Q, GE M F, et al. Design of flow-phase single-molecule immunoassay detection system[J]. Chinese Optics, 2025, 18(5): 1055-1065. (in Chinese). doi: 10.37188/CO.2025-0045 [8] EGLI A. ChatGPT, GPT-4, and other large language models: the next revolution for clinical microbiology?[J]. Clinical Infectious Diseases, 2023, 77(9): 1322-1328. doi: 10.1093/cid/ciad407 [9] ZHU SH B, HONG J K, WANG T. Horizontal gene transfer is predicted to overcome the diversity limit of competing microbial species[J]. Nature Communications, 2024, 15(1): 800. doi: 10.1038/s41467-024-45154-w [10] BELIBASAKIS G N, BELSTROM D, EICK S, et al. Periodontal microbiology and microbial etiology of periodontal diseases: historical concepts and contemporary perspectives[J]. Periodontology 2000, 2023, 92: 11-27. [11] BARON E J, MILLER J M, WEINSTEIN M P, et al. A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2013 recommendations by the Infectious Diseases Society of America (IDSA) and the American Society for Microbiology (ASM)[J]. Clinical Infectious Diseases, 2013, 57(4): e22-e121. doi: 10.1093/cid/cit278 [12] HUANG C, GUO F G, WANG H, et al. An automated system for interrogating the evolution of microbial endosymbiosis[J]. Lab on A Chip, 2023, 23(4): 671-683. doi: 10.1039/D2LC00602B [13] HUANG Y M, SHETH R U, ZHAO SH J, et al. High-throughput microbial culturomics using automation and machine learning[J]. Nature Biotechnology, 2023, 41(10): 1424-1433. doi: 10.1038/s41587-023-01674-2 [14] SAUER S, KLIEM M. Mass spectrometry tools for the classification and identification of bacteria[J]. Nature Reviews Microbiology, 2010, 8(1): 74-82. doi: 10.1038/nrmicro2243 [15] WANG G C, ZHAO B X, ZHENG K F, et al. Research on high-precision gas concentration inversion based on adaptive infrared multi-band joint spectral analysis[J]. Chinese Optics, 2024, 17(6): 1340-1350. (in Chinese). doi: 10.37188/CO.2024-0071 [16] DEKTER H E, ORELIO C C, MORSINK M C, et al. Antimicrobial susceptibility testing of Gram-positive and -negative bacterial isolates directly from spiked blood culture media with Raman spectroscopy[J]. European Journal of Clinical Microbiology & Infectious Diseases, 2017, 36(1): 81-89. [17] NI P X, DING X, ZHANG Y X, et al. Rapid detection and identification of infectious pathogens based on high-throughput sequencing[J]. Chinese Medical Journal, 2015, 128(7): 877-883. doi: 10.4103/0366-6999.154281 [18] CHEN W B, ZHANG CH G. An automated bacterial colony counting and classification system[J]. Information Systems Frontiers, 2009, 11(4): 349-368. doi: 10.1007/s10796-009-9149-0 [19] WANG B X, SONG Y S, DONG X N. Indistinguishable points attention-aware network for infrared small object detection[J]. Chinese Optics, 2024, 17(3): 538-547. (in Chinese). doi: 10.37188/CO.2023-0178 [20] EDUARDO L G, RAMIREZ B S, MARIBEL C F, et al. Low accuracy of the McFarland method for estimation of bacterial populations[J]. African Journal of Microbiology Research, 2018, 12(31): 736-740. doi: 10.5897/AJMR2018.8893 [21] BOIKO D A, MACKNIGHT R, KLINE B, et al. Autonomous chemical research with large language models[J]. Nature, 2023, 624(7992): 570-578. doi: 10.1038/s41586-023-06792-0 [22] SZYMANSKI N J, RENDY B, FEI Y X, et al. An autonomous laboratory for the accelerated synthesis of novel materials[J]. Nature, 2023, 624(7990): 86-91. doi: 10.1038/s41586-023-06734-w [23] BRAN A M, COX S, SCHILTER O, et al. Augmenting large language models with chemistry tools[J]. Nature Machine Intelligence, 2024, 6(5): 525-535. doi: 10.1038/s42256-024-00832-8 [24] XIONG Y, YAO L P, LIN J L, et al. Artificial intelligence links CT images to pathologic features and survival outcomes of renal masses[J]. Nature Communications, 2025, 16(1): 1425. doi: 10.1038/s41467-025-56784-z [25] CALVARESE M, CORBETTA E, CONTRERAS J, et al. Endomicroscopic AI-driven morphochemical imaging and fs-laser ablation for selective tumor identification and selective tissue removal[J]. Science Advances, 2024, 10(50): eado9721. doi: 10.1126/sciadv.ado9721 [26] BAI DX, LI G F, JIANG D, et al. Surface defect detection methods for industrial products with imbalanced samples: a review of progress in the 2020s[J]. Engineering Applications of Artificial Intelligence, 2024, 130: 107697. doi: 10.1016/j.engappai.2023.107697 [27] KOU P, ZHI S F, CHENG Y, et al. Detection of elliptical components in adaptive optical image of space target[J]. Chinese Optics, 2022, 15(3): 454-463. (in Chinese). doi: 10.37188/CO.2021-0208 [28] HUANG Y H, ZHENG W ZH, ZHANG B R, et al. SelfOcc: self-supervised vision-based 3D occupancy prediction[C]. Proceedings of 2024 IEEE/CVF Conference on Computer Vision and Pattern Recognition, IEEE, 2024: 19946-19956. [29] SHAO H, WANG L T, CHEN R B, et al. ReasonNet: end-to-end driving with temporal and global reasoning[C]. Proceedings of 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition, IEEE, 2023: 13723-13733. [30] BURNS B L, RHOADS D D, MISRA A. The use of machine learning for image analysis artificial intelligence in clinical microbiology[J]. Journal of Clinical Microbiology, 2023, 61(9): e02336-21. [31] Khasim S, Ghosh H, Rahat IS, et al. Deciphering Microorganisms through Intelligent Image Recognition: Machine Learning and Deep Learning Approaches, Challenges, and Advancements[J]. EAI Endorsed Transactions on Internet of Things, 2024, 10(1): 1-17. [32] WANG C Y, BOCHKOVSKIY A, LIAO H Y M. YOLOv7: trainable bag-of-freebies sets new state-of-the-art for real-time object detectors[C]. Proceedings of 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition, IEEE, 2023: 7464-7475. [33] ZAND M, ETEMAD A, GREENSPAN M. Objectbox: from centers to boxes for anchor-free object detection[C]. Proceedings of the 17th European Conference on Computer Vision, Springer, 2022: 390-406. [34] ZHAO Y, LV W Y, XU SH L, et al. Detrs beat YOLOs on real-time object detection[C]. Proceedings of 2024 IEEE/CVF Conference on Computer Vision and Pattern Recognition, IEEE, 2024: 16965-16974. -
-
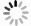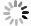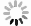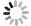




 下载:
下载: