Research progress of single molecule biological detection methods and applications
-
摘要:
单分子生物检测技术是通过了解单分子层面上各生物分子间的动态特性,以发掘生物分子的结构与功能的高效技术。该技术的优势在于能够在单个分子上探测自由能的异质性,这是传统方法无法实现的。利用这一性能,研究人员可以解决复杂生物系统、多相催化、生物分子相互作用、酶系统和构象变化等长期存在的问题。在医疗检测方面,检测单个分子的具体信息或它们与生物因子的相互作用,不仅对癌症等各种疾病的早期诊断和治疗至关重要,而且在实时检测和精准医疗方面具有巨大的潜力。利用单分子生物检测高特异性和高精度的优势,实现对分子群中单个生物分子的实时检测,且可与阵列高通量分析相结合对临床样本进行精确诊断。本文简要介绍了单分子检测原理及其在生物传感方面的应用,在此基础上,重点概述了检测方法及相关应用,最后探讨了该研究方向的前景与发展方向。
Abstract:Single molecule biological detection technology is an efficient technology to understand the dynamic characteristics of various biomolecules at the single molecule level and explore their structure and function. The advantage of this technology is that it can detect the heterogeneity of free energy on a single molecule, which is beyond the traditional methods. Therefore, researchers use it to solve long-standing problems in complex biological systems, heterogeneous catalysis, biomolecular interactions, enzyme systems and conformational changes. In terms of medical detection, detecting specific information about single molecules or their interactions with biological factors is not only crucial for the early diagnosis and treatment of various diseases such as cancer, but also has great potential for real-time detection and precision medicine. The advantages of high specificity and high precision of single-molecule bioassays are used to real-time detection of single biomolecules in molecular populations, and can be combined with multiple high-throughput analysis for the precise diagnosis of clinical samples. In this paper, the principle of single molecule detection and the application of biosensing are introduced, and the detection methods and related applications are summarized. Finally, the prospect and development direction of this research direction are discussed.
-
Key words:
- single molecule biological detection /
- nucleic acid /
- nanopore /
- protein /
- heterogeneity
-
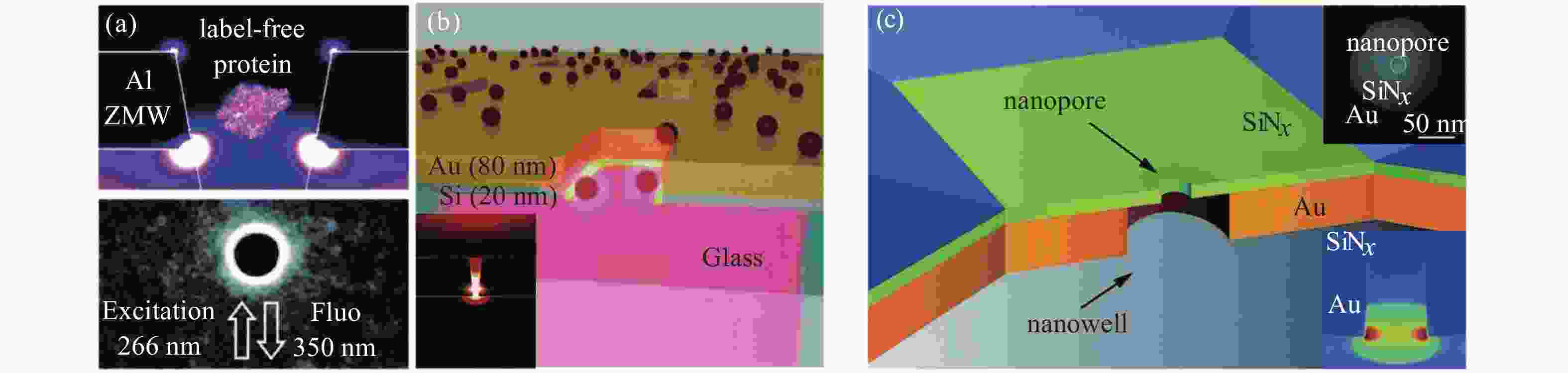
图 2 不同材料架构的零模波导(ZMW)等离子体纳米孔[20]。(a)深紫外等离子体增强Al ZMW单蛋白自身荧光。(b)用于增强单分子探测的Au-Si零模混合波导。(c)等离子体纳米孔器件结构增强单分子荧光检测,该结构由金膜制备的纳米孔和独立式氮化硅膜制备的纳米孔组成。
Figure 2. Various material framework of zero-mode waveguide (ZMW) plasma nanopore[20]. (a)The autofluorescence of Al ZMW monoprotein was enhanced by deep ultraviolet plasma. (b) Au-Si zero-mode hybrid waveguides for enhanced single-molecule detection. (c)The plasma nanopore device structure enhances single-molecule fluorescence detection, which consists of nanopore prepared by gold film and nanopore prepared by independent silicon nitride film.
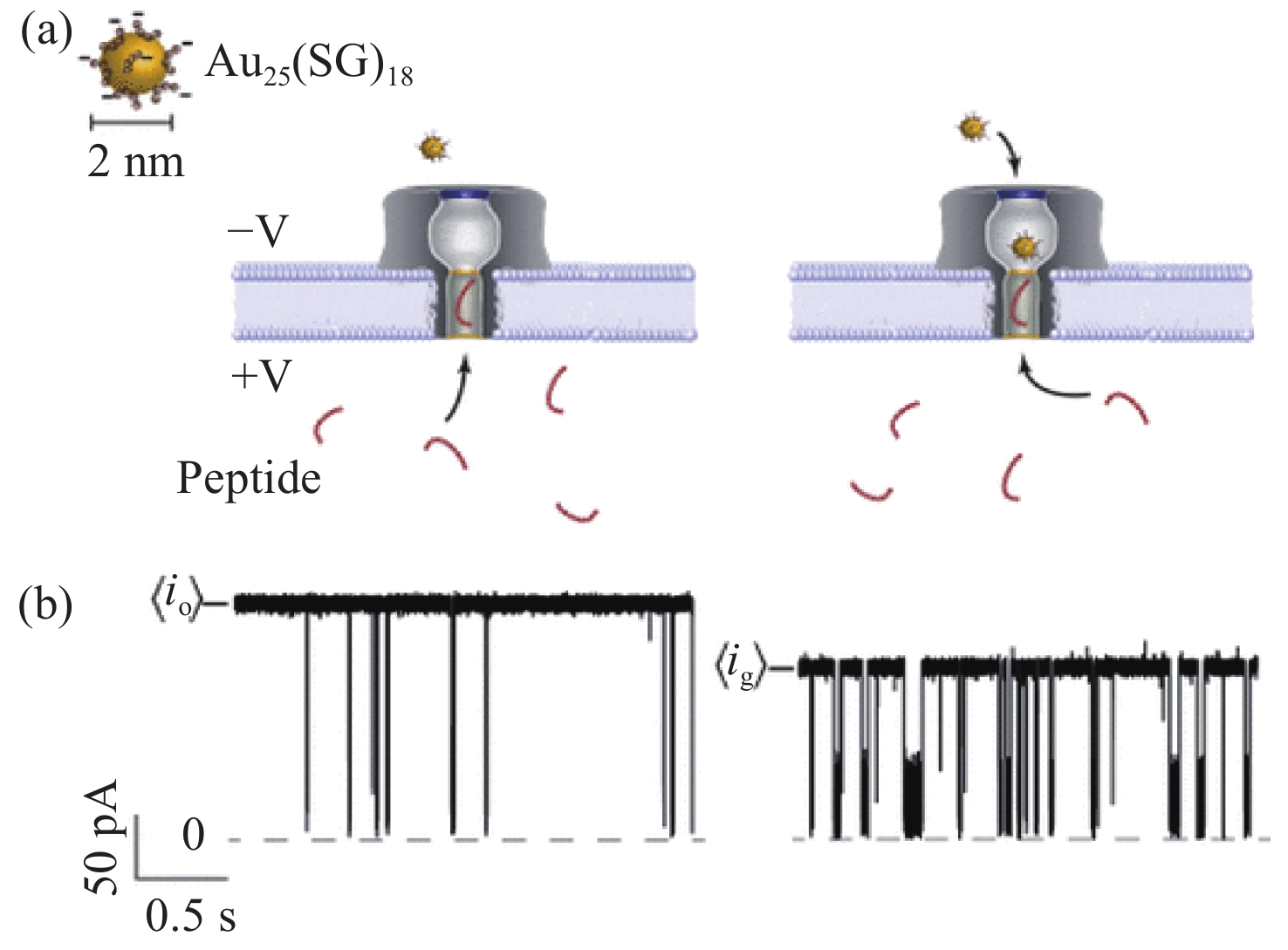
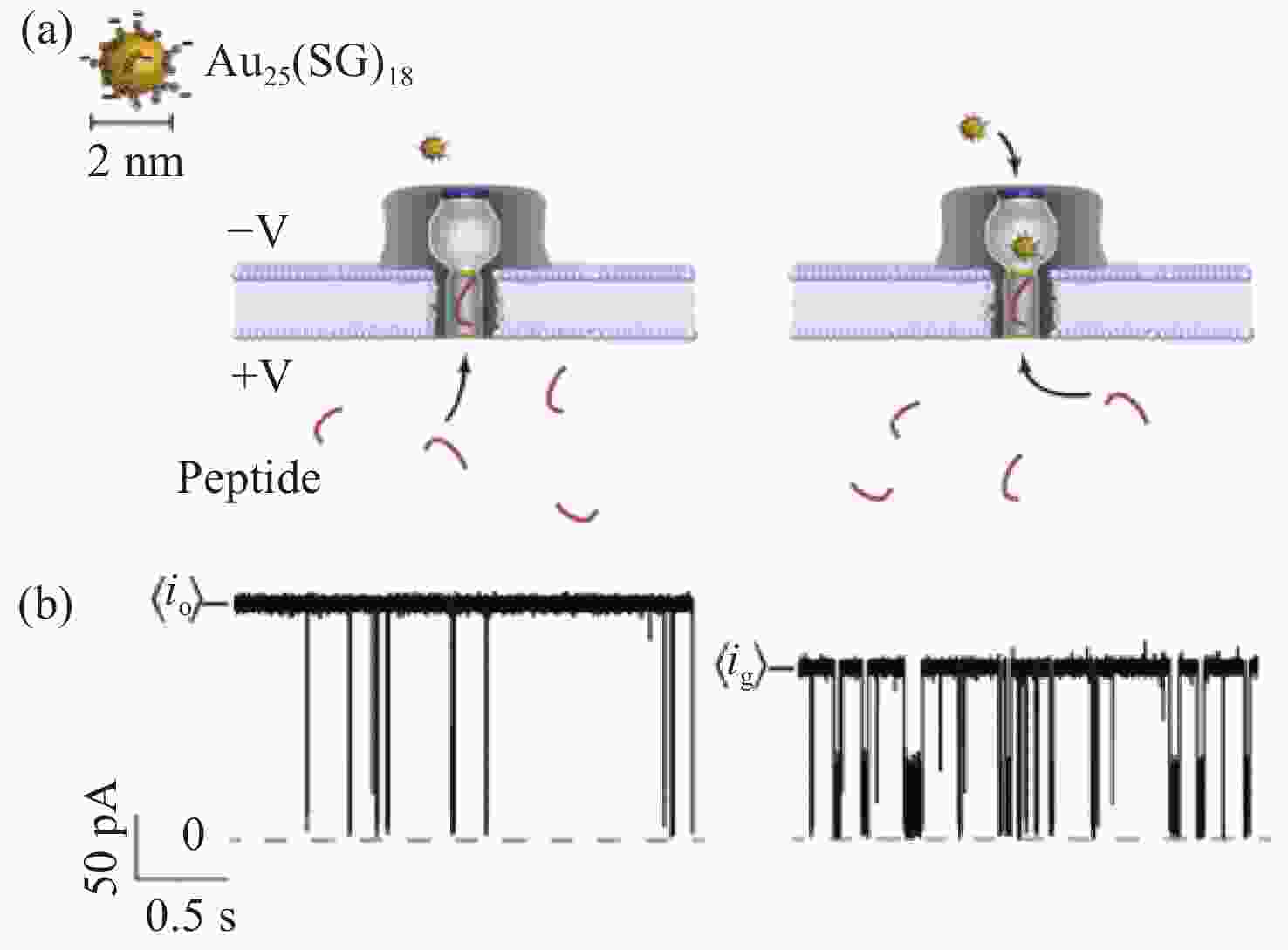
图 3 通过野生型气溶胶膜通道运输C-A3和mC-A3[33]。(a)气溶胶纳米孔模型。气溶素(灰色)嵌入脂质双层膜(深蓝色),核苷酸(红色)放置在孔的入口;(b)含有甲基胞嘧啶和胞嘧啶的甲基化和非甲基化寡核苷酸结构。红色部分为添加的甲基部分
Figure 3. Transporting C-A3 and mC-A3 through a wild-type aerolysin membrane channel[33]. (a) All-atom model of the full-length aerolysin nanopore system. Aerolysin (gray) was inserted into a lipid bilayer membrane (dark blue), while nucleotides (red) were placed at the entrance of the pore. (b) Structure of methylated and unmethylated oligonucleotides containing methylcytosine and cytosine, respectively. The only difference between them was the addition of a methyl group which is marked in red
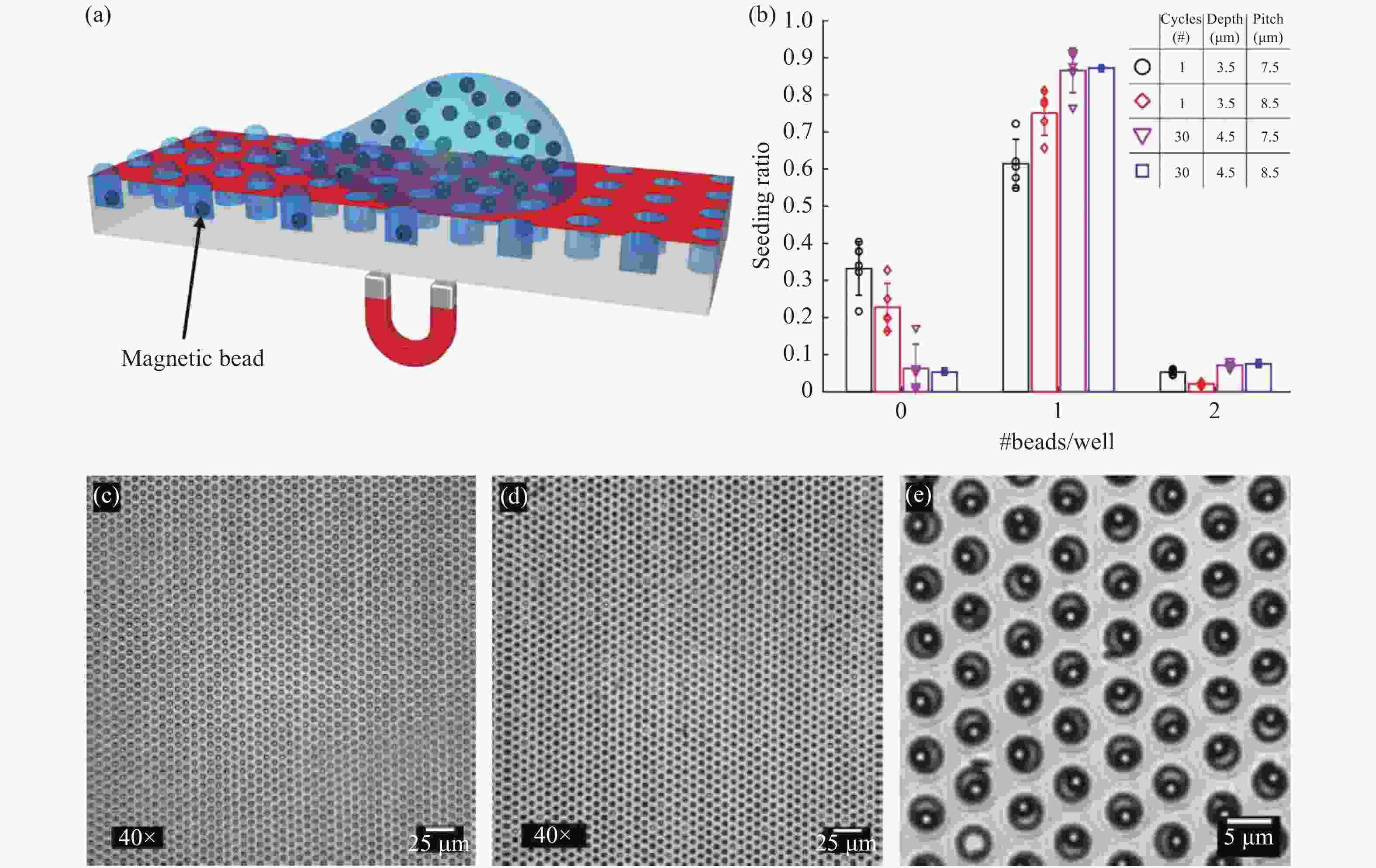
图 4 微磁珠加载示意图[35]。(a)使用磁力和亲-疏水微孔阵列高效加载微磁珠。(b)直径、间距和深度不同的微孔阵列在多次循环下的加载率。(c、d)40倍显微镜下加载前后的亮场显微图像。(e)100倍显微镜下加载后的亮场显微图像
Figure 4. Magnetic bead seeding[35]. (a) Schematic of the magnetic bead seeding on HIH microwell arrays. (b) The bead distribution in arrays with varying well diameters, array pitches and well depths, for multiple-seeding cycles. (c,d) ×40 magnification bright field microscopy image of a microwell array before and after magnetic bead seeding, respectively. (e) ×100 magnification bright field microscopy image of a microwell array after seeding
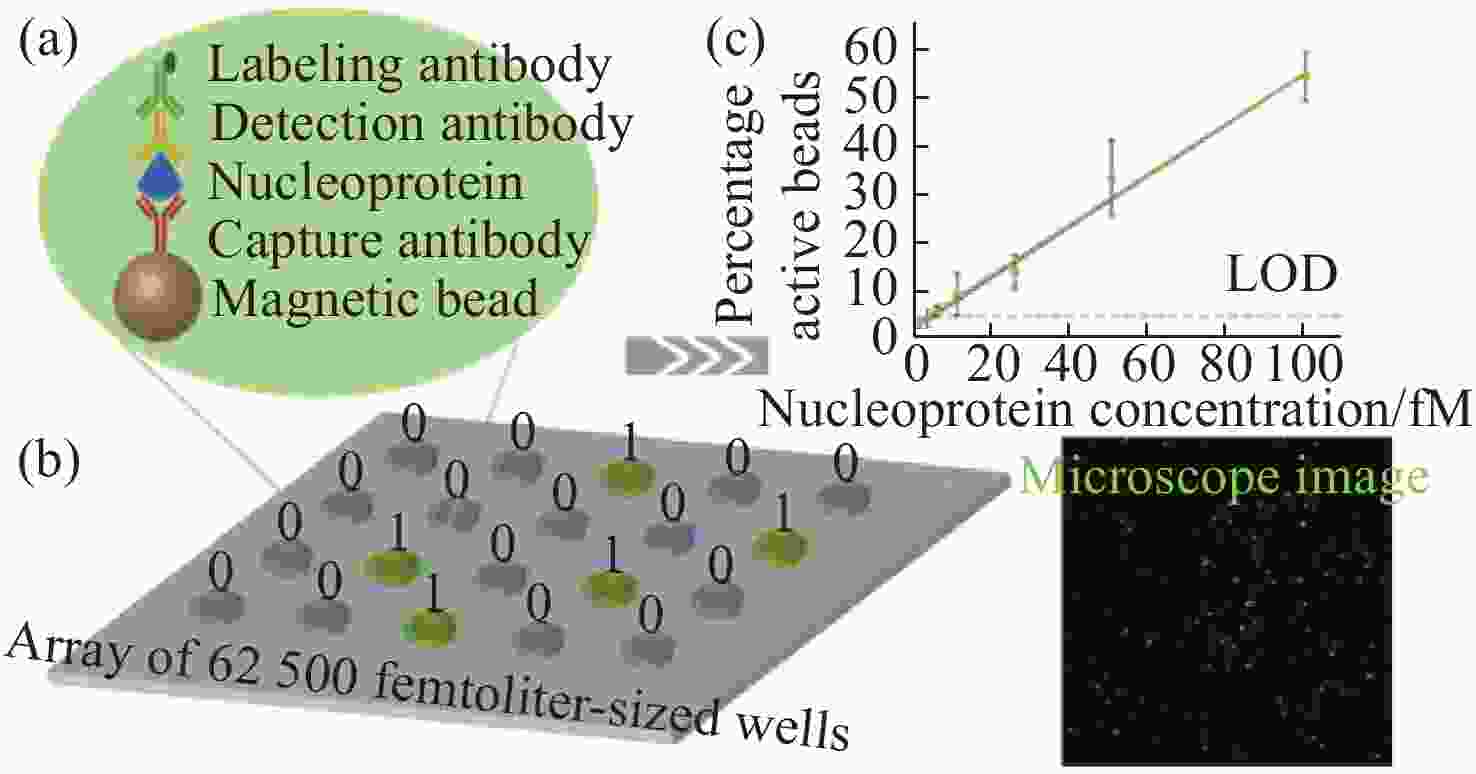
图 5 数字ELISA示意图[37]。(a)抗体结合微珠捕获单个目标生物分子,然后由另一个与标记抗体结合的抗体检测。将微珠装入飞升微孔阵列中用于分离和检测单个分子。(b)产生单分子信号的飞升微孔阵列的一部分荧光图像。大多数飞升体积微孔含有一颗微珠,但这些珠中只有一小部分具有催化酶活性,表明是一种单一的结合蛋白。(c)蛋白质在本体溶液中的浓度与结合蛋白质分子的微珠的百分比关系
Figure 5. Schematic representation of digital ELISA[37]. (a) Antibody-coated beads captures the single target biomolecules, which are then detected by another antibody conjugated with a labeled antibody. Loading of beads into femtoliter well arrays for isolation and detection of single molecules. (b) Fluorescence image of a small section of the femtoliter well array after signals from single molecules are generated. While the majority of femtoliter chambers contain a bead from the assay, only a fraction of those beads possesses catalytic enzyme activity, indicative of a single, bound protein. (c) The concentration of protein in the bulk solution is correlated to the percentage of beads that have bound a protein molecule
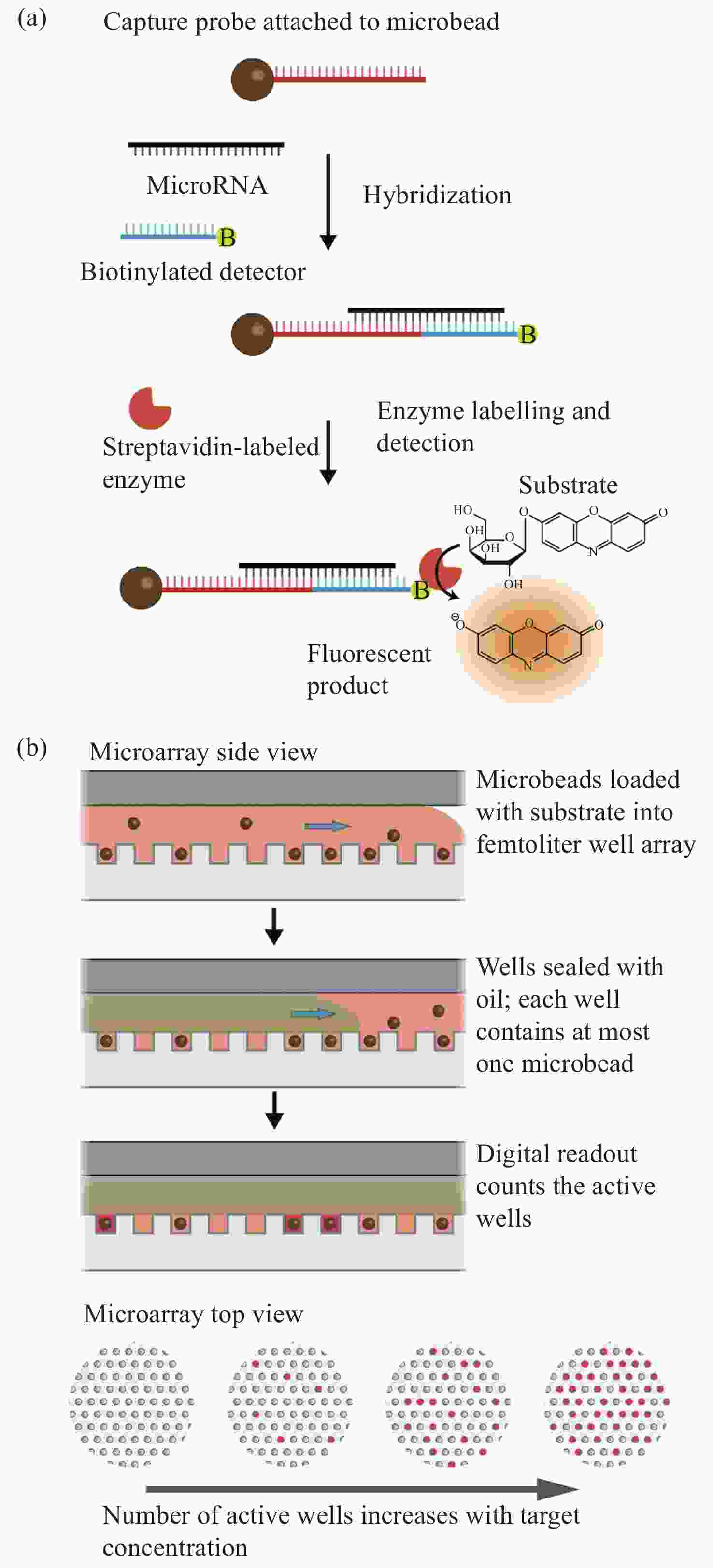
图 6 数字式单分子阵列检测技术进行MicroRNA检测[43]。(a)单个miRNA分子通过与生物素化的探测器探针杂交到探针化的顺磁珠中被捕获。随后加入链霉亲和素结合酶来标记捕获的miRNA复合物,以便在与荧光酶底物孵育时产生荧光信号。(b)单个微珠与荧光衬底一起装入飞升微孔阵列,随后用油密封。然后将孔上荧光的数量作为目标miRNA浓度的数字读数
Figure 6. MicroRNA detection with digital single molecule detection technology[43]. (a) Individual miRNA molecules are captured by hybridization to probecoated paramagnetic beads, along with biotinylated detector probe. The streptavidin-conjugated enzyme is subsequently added to label the captured miRNA complex, to allow generation of a fluorescent signal upon incubation with a fluorogenic enzyme substrate. (b) Individual beads are loaded along with fluorogenic substrate into a femtoliter microwell array, which is subsequently sealed with oil. The number of fluorescent “on” wells is then counted as a digital readout of the target miRNA concentration
表 1 文中不同单分子生物检测方法的优缺点对比
Table 1. Advantages and disadvantages of various methods of single molecule biological detection
Single molecule methods Main detection molecules Main advantages Main disadvantages Nanopore Long and short chains of DNA and RNA, proteins, and polypeptide Required low sample volume; compact and simple;
allows label free detectionProne to error; high cost SERS Virus, protein, biomarkers High sensitivity; high selectivity Nonspecific binding with interfering molecule can give false signals Digital single molecule Nucleic acid, protein, small molecule High specificity; quantitative; wide dynamic range Limited multiplexing capabilities; high cost WGM Virus, Protein conformation etc. High sensitivity; easy to manufacture; low cost Low specificity, limited to frequency
and wavelengthCRISPR Nucleic acid Low cost; high efficiency Not suitable for multiple analyte detection, required sample pretreatment -
[1] HALL C E. Method for the observation of macromolecules with the electron microscope illustrated with micrographs of DNA[J]. The Journal of Biophysical and Biochemical Cytology, 1956, 2(5): 625-628. doi: 10.1083/jcb.2.5.625 [2] ROTMAN B. Measurement of activity of single molecules of β -D-galactosidase[J]. Proceedings of the National Academy of Sciences of the United States of America, 1961, 47(12): 1981-1991. doi: 10.1073/pnas.47.12.1981 [3] MILLER H, ZHOU ZH K, SHEPHERD J, et al. Single-molecule techniques in biophysics: a review of the progress in methods and applications[J]. Reports on Progress in Physics, 2018, 81(2): 024601. doi: 10.1088/1361-6633/aa8a02 [4] HIRSCHFELD T. Optical microscopic observation of single small molecules[J]. Applied Optics, 1976, 15(12): 2965-2966. doi: 10.1364/AO.15.002965 [5] GELLES J, SCHNAPP B J, SHEETZ M P. Tracking kinesin-driven movements with nanometre-scale precision[J]. Nature, 1988, 331(6155): 450-453. doi: 10.1038/331450a0 [6] ORRIT M, BERNARD J. Single pentacene molecules detected by fluorescence excitation in a p-terphenyl crystal[J]. Physical Review Letters, 1990, 65(21): 2716-2719. doi: 10.1103/PhysRevLett.65.2716 [7] KNEIPP K, WANG Y, KNEIPP H, et al. Single molecule detection using surface-enhanced Raman scattering (SERS)[J]. Physical Review Letters, 1997, 78(9): 1667-1670. doi: 10.1103/PhysRevLett.78.1667 [8] LU H P, XUN L, XIE X S. Single molecule enzymatic dynamics[J]. Science, 1998, 282(5395): 1877-1882. [9] VOGELSTEIN B, KINZLER K W. Digital PCR[J]. Proceedings of the National Academy of Sciences of the United States of America, 1999, 96(16): 9236-9241. doi: 10.1073/pnas.96.16.9236 [10] KOREN S, SCHATZ M C, WALENZ B P, et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads[J]. Nature Biotechnology, 2012, 30(7): 693-700. doi: 10.1038/nbt.2280 [11] GOOTENBERG J S, ABUDAYYEH O O, LEE J W, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2[J]. Science, 2017, 356(6336): 438-442. doi: 10.1126/science.aam9321 [12] FUXREITER M, VENDRUSCOLO M. Generic nature of the condensed states of proteins[J]. Nature Cell Biology, 2021, 23(6): 587-594. doi: 10.1038/s41556-021-00697-8 [13] WANG Y Q, GUAN X Y, ZHANG S Y, et al. Structural-profiling of low molecular weight RNAs by nanopore trapping/translocation using Mycobacterium smegmatis porin A[J]. Nature Communications, 2021, 12(1): 3368. doi: 10.1038/s41467-021-23764-y [14] RUAN J B, XIA SH Y, LIU X, et al. Cryo-EM structure of the gasdermin A3 membrane pore[J]. Nature, 2018, 557(7703): 62-67. doi: 10.1038/s41586-018-0058-6 [15] WANG J Z, HU M J, WANG J, et al. Reconstitution and structure of a plant NLR resistosome conferring immunity[J]. Science, 2019, 364(6435): eaav5870. doi: 10.1126/science.aav5870 [16] LANGECKER M, ARNAUT V, MARTIN T G, et al. Synthetic lipid membrane channels formed by designed DNA nanostructures[J]. Science, 2012, 338(6109): 932-936. doi: 10.1126/science.1225624 [17] KASIANOWICZ J J, BRANDIN E, BRANTON D. Characterization of individual polynucleotide molecules using a membrane channel[J]. Proceedings of the National Academy of Sciences of the United States of America, 1996, 93(24): 13770-13773. doi: 10.1073/pnas.93.24.13770 [18] CHAVIS A E, BRADY K T, HATMAKER G A, et al. Single molecule nanopore spectrometry for peptide detection[J]. ACS Sensors, 2017, 2(9): 1319-1328. doi: 10.1021/acssensors.7b00362 [19] RAHMAN M M, SAMPAD M J N, HAWKINS A, et al. Recent advances in integrated solid-state nanopore sensors[J]. Lab on a Chip, 2021, 21(16): 3030-3052. doi: 10.1039/D1LC00294E [20] MACCAFERRIN, BARBILLON G, KOYAAN, et al. Recent advances in plasmonic nanocavities for single-molecule spectroscopy[J]. Nanoscale Advance, 2021, 3(3): 633-642. [21] ASSAD O N, GILBOA T, SPITZBERG J, et al. Light-enhancing plasmonic-nanopore biosensor for superior single-molecule detection[J]. Advanced Materials, 2017, 29(9): 1605442. doi: 10.1002/adma.201605442 [22] BUCHFINK B, REUTER K, DROST H G. Sensitive protein alignments at tree-of-life scale using DIAMOND[J]. Nature Methods, 2021, 18(4): 366-368. doi: 10.1038/s41592-021-01101-x [23] BELLENGUEZ C, KÜÇÜKALI F, JANSEN I E, et al. New insights into the genetic etiology of Alzheimer's disease and related dementias[J]. Nature Genetics, 2022, 54(4): 412-436. doi: 10.1038/s41588-022-01024-z [24] DEVESON I W, GONG B SH, LAI K, et al. Evaluating the analytical validity of circulating tumor DNA sequencing assays for precision oncology[J]. Nature Biotechnology, 2021, 39(9): 1115-1128. doi: 10.1038/s41587-021-00857-z [25] BRINKERHOFF H, KANG A S W, LIU J Q, et al.. Infinite re-reading of single proteins at single-amino-acid resolution using nanopore sequencing[J]. BioRxiv, 2021, doi: 10.1126/science.abl4381. [26] SEN P, GUPTA M. Single nucleotide detection using bilayer MoS2 nanopores with high efficiency[J]. RSC Advances, 2021, 11(11): 6114-6123. doi: 10.1039/D0RA10222A [27] YAMAZAKI H, HU R, ZHAO Q, et al. Photothermally assisted thinning of silicon nitride membranes for ultrathin asymmetric nanopores[J]. ACS Nano, 2018, 12(12): 12472-12481. doi: 10.1021/acsnano.8b06805 [28] BURCK N, GILBOA T, GADI A, et al. Nanopore identification of single nucleotide mutations in circulating tumor DNA by multiplexed ligation[J]. Clinical Chemistry, 2021, 67(5): 753-762. doi: 10.1093/clinchem/hvaa328 [29] YUAN B, LI SH, YING Y L, et al. The analysis of single cysteine molecules with an aerolysin nanopore[J]. Analyst, 2020, 145(4): 1179-1183. doi: 10.1039/C9AN01965K [30] PIGUET F, OULDALI H, PASTORIZA-GALLEGO M, et al. Identification of single amino acid differences in uniformly charged homopolymeric peptides with aerolysin nanopore[J]. Nature Communications, 2018, 9(1): 966. doi: 10.1038/s41467-018-03418-2 [31] CAO C, CIRAUQUI N, MARCAIDA M J, et al. Single-molecule sensing of peptides and nucleic acids by engineered aerolysin nanopores[J]. Nature communications, 2019, 10(1): 1-11. [32] ZHANG SH L, HUANG G, VERSLOOT R C A, et al. Bottom-up fabrication of a proteasome–nanopore that unravels and processes single proteins[J]. Nature Chemistry, 2021, 13(12): 1192-1199. doi: 10.1038/s41557-021-00824-w [33] LI M Y, YING Y L, YU J, et al. Revisiting the origin of nanopore current blockage for volume difference sensing at the atomic level[J]. JACS Au, 2021, 1(7): 967-976. doi: 10.1021/jacsau.1c00109 [34] LEIRS K, KUMAR P T, DECROP D, et al. Bioassay development for ultrasensitive detection of influenza a nucleoprotein using digital ELISA[J]. Analytical Chemistry, 2016, 88(17): 8450-8458. doi: 10.1021/acs.analchem.6b00502 [35] SHAFAGH R Z, DECROP D, VEN K, et al. Reaction injection molding of hydrophilic-in-hydrophobic femtolitre-well arrays[J]. Microsystems &Nanoengineering, 2019, 5: 25. [36] RISSIN D M, WALT D R. Digital readout of target binding with attomole detection limits via enzyme amplification in femtoliter arrays[J]. Journal of the American Chemical Society, 2006, 128(19): 6286-6287. doi: 10.1021/ja058425e [37] RISSIN D M, KAN C W, CAMPBELL T G, et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations[J]. Nature Biotechnology, 2010, 28(6): 595-599. doi: 10.1038/nbt.1641 [38] LEE J, CRAMPTON K T, TALLARIDA N, et al. Visualizing vibrational normal modes of a single molecule with atomically confined light[J]. Nature, 2019, 568(7750): 78-82. doi: 10.1038/s41586-019-1059-9 [39] YERA H, OK V, KUET F L K, et al. PCR and culture for diagnosis of Acanthamoeba keratitis[J]. British Journal of Ophthalmology, 2021, 105(9): 1302-1306. doi: 10.1136/bjophthalmol-2020-316730 [40] HINDSON B J, NESS K D, MASQUELIER D A, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number[J]. Analytical Chemistry, 2011, 83(22): 8604-8610. doi: 10.1021/ac202028g [41] CHEN Y J, QIAN CH, LIU CH ZH, et al. Nucleic acid amplification free biosensors for pathogen detection[J]. Biosensors and Bioelectronics, 2020, 153: 112049. doi: 10.1016/j.bios.2020.112049 [42] GINES G, MENEZES R, NARA K, et al. Isothermal digital detection of microRNAs using background-free molecular circuit[J]. Science Advances, 2020, 6(4): eaay5952. doi: 10.1126/sciadv.aay5952 [43] COHEN L, HARTMAN M R, AMARDEY-WELLINGTON A, et al. Digital direct detection of microRNAs using single molecule arrays[J]. Nucleic Acids Research, 2017, 45(14): e137. doi: 10.1093/nar/gkx542 [44] ASHTON N J, LEUZY A, KARIKARI T K, et al. The validation status of blood biomarkers of amyloid and phospho-tau assessed with the 5-phase development framework for AD biomarkers[J]. European Journal of Nuclear Medicine and Molecular Imaging, 2021, 48(7): 2140-2156. doi: 10.1007/s00259-021-05253-y [45] GILL J, LATOUR L, DIAZ-ARRASTIA R, et al. Glial fibrillary acidic protein elevations relate to neuroimaging abnormalities after mild TBI[J]. Neurology, 2018, 91(15): e1385-e1389. doi: 10.1212/WNL.0000000000006321 [46] THELIN E, AL NIMER F, FROSTELL A, et al. A serum protein biomarker panel improves outcome prediction in human traumatic brain injury[J]. Journal of Neurotrauma, 2019, 36(20): 2850-2862. doi: 10.1089/neu.2019.6375 [47] PÉREZ-RUIZ E, DECROP D, VEN K, et al. Digital ELISA for the quantification of attomolar concentrations of Alzheimer's disease biomarker protein Tau in biological samples[J]. Analytica Chimica Acta, 2018, 1015: 74-81. doi: 10.1016/j.aca.2018.02.011 [48] DINH T L, NGAN K C, SHOEMAKER C B, et al. Rapid and ultrasensitive detection of botulinum neurotoxin serotype A1 in human serum and urine using single-molecule array method[J]. Forensic Toxicology, 2017, 35(1): 179-184. doi: 10.1007/s11419-016-0336-7 [49] WANG X, COHEN L, WANG J, et al. Competitive immunoassays for the detection of small molecules using single molecule arrays[J]. Journal of the American Chemical Society, 2018, 140(51): 18132-18139. doi: 10.1021/jacs.8b11185 [50] KIM E, BAASKE M D, VOLLMER F. Towards next-generation label-free biosensors: recent advances in whispering gallery mode sensors[J]. Lab on a Chip, 2017, 17(7): 1190-1205. doi: 10.1039/C6LC01595F [51] SUBRAMANIAN S, VINCENT S, VOLLMER F. Effective linewidth shifts in single-molecule detection using optical whispering gallery modes[J]. Applied Physics Letters, 2020, 117(15): 151106. doi: 10.1063/5.0028113 [52] SANTIAGO-CORDOBA M A, CETINKAYA M, BORISKINA S V, et al. Ultrasensitive detection of a protein by optical trapping in a photonic-plasmonic microcavity[J]. Journal of Biophotonics, 2012, 5(8-9): 629-638. doi: 10.1002/jbio.201200040 [53] YU W Y, JIANG W C, LIN Q, et al. Cavity optomechanical spring sensing of single molecules[J]. Nature Communications, 2016, 7: 12311. doi: 10.1038/ncomms12311 [54] BAILEY R C, WASHBURN A L, QAVI A J, et al. A robust silicon photonic platform for multiparameter biological analysis[J]. Proceedings of SPIE, 2009, 7220: 72200N. doi: 10.1117/12.809819 [55] SHI H X, CUI J J, SULEMANA H, et al. Protein detection based on rolling circle amplification sensors[J]. Luminescence, 2021, 36(4): 842-848. doi: 10.1002/bio.4017 [56] NITU F R, SAVU L, MURARU S, et al. Label-free homogeneous microRNA detection in cell culture medium based on graphene oxide and specific fluorescence quenching[J]. Nanomaterials, 2021, 11(2): 368. doi: 10.3390/nano11020368 [57] VOLLMER F, ARNOLD S, KENG D. Single virus detection from the reactive shift of a whispering-gallery mode[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(52): 20701-20704. doi: 10.1073/pnas.0808988106 [58] CARDENOSA-RUBIO M C, ROBISON H M, BAILEY R C. Recent advances in environmental and clinical analysis using microring resonator–based sensors[J]. Current Opinion in Environmental Science &Health, 2019, 10: 38-46. [59] SHAO L B, JIANG X F, YU X CH, et al. Detection of single nanoparticles and lentiviruses using microcavity resonance broadening[J]. Advanced Materials, 2013, 25(39): 5616-5620. doi: 10.1002/adma201302572 [60] DOMINGUEZ I, DEL VILLAR I, FUENTES O, et al. Dually nanocoated planar waveguides towards multi-parameter sensing[J]. Scientific Reports, 2021, 11(1): 3669. doi: 10.1038/s41598-021-83324-8 [61] COGNETTI J S, STEINER D J, ABEDIN M, et al. Disposable photonics for cost-effective clinical bioassays: application to COVID-19 antibody testing[J]. Lab on a Chip, 2021, 21(15): 2913-2921. doi: 10.1039/D1LC00369K [62] ROBISON H M, ESCALANTE P, VALERA E, et al. Precision immunoprofiling to reveal diagnostic signatures for latent tuberculosis infection and reactivation risk stratification[J]. Integrative Biology, 2019, 11(1): 16-25. doi: 10.1093/intbio/zyz001 [63] YU X CH, TANG SH J, LIU W J, et al. Single-molecule optofluidic microsensor with interface whispering gallery modes[J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(6): e2108678119. doi: 10.1073/pnas.2108678119 [64] KIM E, BAASKE M D, VOLLMER F. In situ observation of single-molecule surface reactions from low to high affinities[J]. Advanced Materials, 2016, 28(45): 9941-9948. doi: 10.1002/adma.201603153 [65] KIM E, BAASKE M D, SCHULDES I, et al. Label-free optical detection of single enzyme-reactant reactions and associated conformational changes[J]. Science Advances, 2017, 3(3): e1603044. doi: 10.1126/sciadv.1603044 [66] SUBRAMANIAN S, JONES H B L, FRUSTACI S, et al. Sensing enzyme activation heat capacity at the single-molecule level using gold-nanorod-based optical whispering gallery modes[J]. ACS Applied Nano Materials, 2021, 4(5): 4576-4583. doi: 10.1021/acsanm.1c00176 [67] AMBARTSUMYAN O, GRIBANYOV D, KUKUSHKIN V, et al. SERS-based biosensors for virus determination with oligonucleotides as recognition elements[J]. International Journal of Molecular Sciences, 2020, 21(9): 3373. doi: 10.3390/ijms21093373 [68] FLEISCHMANN M, HENDRA P J, MCQUILLAN A J. Raman spectra of pyridine adsorbed at a silver electrode[J]. Chemical Physics Letters, 1974, 26(2): 163-166. doi: 10.1016/0009-2614(74)85388-1 [69] LI W Y, CAMARGO P H C, LU X M, et al. Dimers of silver nanospheres: facile synthesis and their use as hot spots for surface-enhanced Raman scattering[J]. Nano Letters, 2009, 9(1): 485-490. doi: 10.1021/nl803621x [70] BLACKIE E J, LE RU E C, ETCHEGOIN P G. Single-molecule surface-enhanced Raman spectroscopy of nonresonant molecules[J]. Journal of the American Chemical Society, 2009, 131(40): 14466-14472. doi: 10.1021/ja905319w [71] LINDQUIST N C, DE ALBUQUERQUE C D L, SOBRAL-FILHO R G, et al. High-speed imaging of surface-enhanced Raman scattering fluctuations from individual nanoparticles[J]. Nature Nanotechnology, 2019, 14(10): 981-987. doi: 10.1038/s41565-019-0535-6 [72] LI ZH Y. Mesoscopic and microscopic strategies for engineering Plasmon-enhanced Raman scattering[J]. Advanced Optical Materials, 2018, 6(16): 1701097. doi: 10.1002/adom.201701097 [73] YAMPOLSKY S, FISHMAN D A, DEY S, et al. Seeing a single molecule vibrate through time-resolved coherent anti-Stokes Raman scattering[J]. Nature Photonics, 2014, 8(8): 650-656. doi: 10.1038/nphoton.2014.143 [74] ZHANG K, LIU Y J, WANG Y N, et al. Direct SERS tracking of a chemical reaction at a single 13 nm gold nanoparticle[J]. Chemical Science, 2019, 10(6): 1741-1745. doi: 10.1039/C8SC04496A [75] NASIR S, MAJEED M I, NAWAZ H, et al. Surface enhanced Raman spectroscopy of RNA samples extracted from blood of hepatitis C patients for quantification of viral loads[J]. Photodiagnosis and Photodynamic Therapy, 2021, 33: 102152. doi: 10.1016/j.pdpdt.2020.102152 [76] CHEN H, PARK S G, CHOI N, et al. SERS imaging-based aptasensor for ultrasensitive and reproducible detection of influenza virus A[J]. Biosensors and Bioelectronics, 2020, 167: 112496. doi: 10.1016/j.bios.2020.112496 [77] CHAUHAN N, SAXENA K, TIKADAR M, et al. Recent advances in the design of biosensors based on novel nanomaterials: an insight[J]. Nanotechnology and Precision Engineering, 2021, 4(4): 045003. doi: 10.1063/10.0006524 [78] NGUYEN H A, JUPIN I, DECORSE P, et al. Assembly of gold nanoparticles using turnip yellow mosaic virus as an in-solution SERS sensor[J]. RSC Advances, 2019, 9(55): 32296-32307. doi: 10.1039/C9RA08015E [79] KUKUSHKIN V I, IVANOV N M, NOVOSELTSEVA A A, et al. Highly sensitive detection of influenza virus with SERS aptasensor[J]. PLoS One, 2019, 14(4): e0216247. doi: 10.1371/journal.pone.0216247 [80] PENG Y S, LIN C L, LONG L, et al. Charge-transfer resonance and electromagnetic enhancement synergistically enabling MXenes with excellent SERS sensitivity for SARS-CoV-2 S protein detection[J]. Nano-Micro Letters, 2021, 13: 52. doi: 10.1007/s40820-020-00565-4 [81] ANTOINE D, MOHAMMADI M, VITT M, et al. Rapid, point-of-care scFv-SERS assay for femtogram level detection of SARS-CoV-2[J]. ACS Sensors, 2022, 7(3): 866-873. doi: 10.1021/acssensors.1c02664 [82] LIU B, ZHENG SH Y, LI H T, et al. Ultrasensitive and facile detection of multiple trace antibiotics with magnetic nanoparticles and core-shell nanostar SERS nanotags[J]. Talanta, 2022, 237: 122955. doi: 10.1016/j.talanta.2021.122955 [83] SHIN H, OH S, KANG D, et al. Protein quantification and imaging by surface-enhanced Raman spectroscopy and similarity analysis[J]. Advanced Science, 2020, 7(11): 1903638. doi: 10.1002/advs.201903638 [84] ZHOU W, TIAN Y F, YIN B CH, et al. Simultaneous surface-enhanced Raman spectroscopy detection of multiplexed microRNA biomarkers[J]. Analytical Chemistry, 2017, 89(11): 6120-6128. doi: 10.1021/acs.analchem.7b00902 [85] PANG Y F, WANG CH G, LU L CH, et al. Dual-SERS biosensor for one-step detection of microRNAs in exosome and residual plasma of blood samples for diagnosing pancreatic cancer[J]. Biosensors and Bioelectronics, 2019, 130: 204-213. doi: 10.1016/j.bios.2019.01.039 [86] NING C F, WANG L Y, TIAN Y F, et al. Multiple and sensitive SERS detection of cancer-related exosomes based on gold–silver bimetallic nanotrepangs[J]. Analyst, 2020, 145(7): 2795-2804. doi: 10.1039/C9AN02180A [87] LI L, LIU CH, CAO X W, et al. Multiplexing determination of cancer-associated biomarkers by surface-enhanced Raman scattering using ordered gold nanohoneycomb arrays[J]. Bioanalysis, 2017, 9(20): 1561-1572. doi: 10.4155/bio-2016-0237 [88] ATANASOV A G, ZOTCHEV S B, DIRSCH V M, et al. Natural products in drug discovery: advances and opportunities[J]. Nature Reviews Drug Discovery, 2021, 20(3): 200-216. doi: 10.1038/s41573-020-00114-z [89] CARTER L J, GARNER L V, SMOOT J W, et al. Assay techniques and test development for COVID-19 diagnosis[J]. ACS Central Science, 2020, 6(5): 591-605. doi: 10.1021/acscentsci.0c00501 [90] DE PUIG H, LEE R A, NAJJAR D, et al. Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants[J]. Science Advances, 2021, 7(32): eabh2944. doi: 10.1126/sciadv.abh2944 [91] MYHRVOLD C, FREIJE C A, GOOTENBERG J S, et al. Field-deployable viral diagnostics using CRISPR-Cas13[J]. Science, 2018, 360(6387): 444-448. doi: 10.1126/science.aas8836 [92] GOOTENBERG J S, ABUDAYYEH O O, KELLNER M J, et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6[J]. Science, 2018, 360(6387): 439-444. doi: 10.1126/science.aaq0179 [93] MAKAROVA K S, WOLF Y I, IRANZO J, et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants[J]. Nature Reviews Microbiology, 2020, 18(2): 67-83. doi: 10.1038/s41579-019-0299-x -




 下载:
下载: