Recent progress on the reconstruction algorithms of structured illumination microscopy
doi: 10.37188/CO.EN.2022-0011
-
摘要:
作为现代超分辨成像技术的早期组成部分,结构照明显微镜(SIM)已经发展了近20年。其近期在活细胞中实现了高达60 nm和564 Hz的最佳时空分辨率组合,但也存在一些源于内在重建过程的缺点。本文综述了SIM技术的最新进展,包括超分辨率(SR)重建算法、性能评估及SIM与其他成像技术的集成,以便为生物学家提供实用指导。
Abstract:As an early component of modern Super-Resolution (SR) imaging technology, Structured Illumination Microscopy (SIM) has been developed for nearly twenty years. With up to ~60 nm wavelengths and 564 Hz frame rates, it has recently achieved an optimal combination of spatiotemporal resolution in live cells. Despite these advantages, SIM also suffers disadvantages, some of which originated from the intrinsic reconstruction process. Here we review recent technical advances in SIM, including SR reconstruction, performance evaluation, and its integration with other technologies to provide a practical guide for biologists.
-
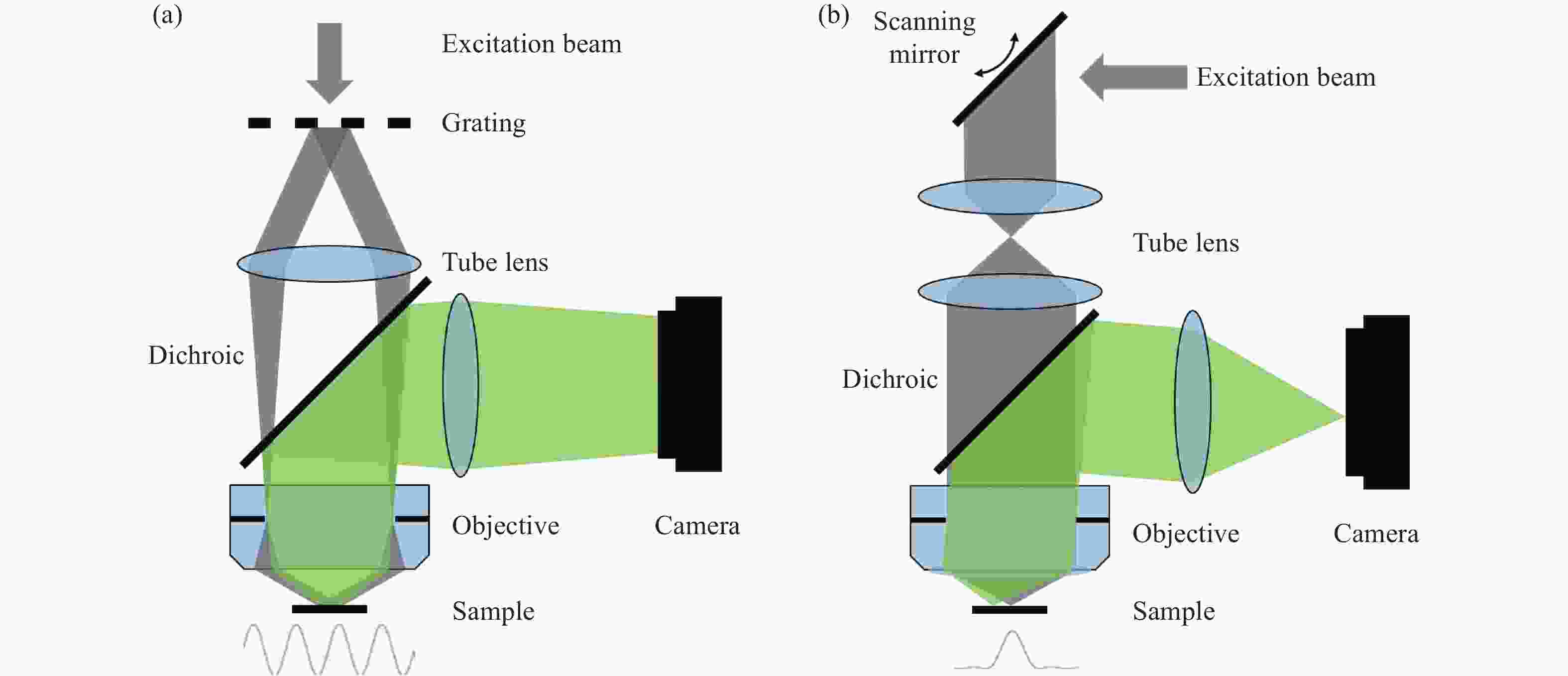
Figure 1. Schematic diagram of structured illumination microscopy. (a) In sinusoidal illumination microscopy, interference between multiple beams (usually generated by a diffraction grating or spatial light modulator) creates a 2D or 3D striped pattern with spatial frequency
${k}_{{\rm{ex}}}$ illuminating on the sample. This pattern shifts the sample's spatial frequency spectrum$ \stackrel{~}{S}\left(k\right) $ to$\stackrel{~}{S}\left(k+{k}_{{\rm{ex}}}\right)$ and$\stackrel{~}{S}\left(k-{k}_{{\rm{ex}}}\right)$ , translating high-frequency SR information into the diffraction-limited detection passband${OTF}_{{\rm{em}}}\left(k\right)$ with the spatial cutoff frequency${k}_{{\rm{em}}}$ . After computational processing, the sample's highest detectable frequency can be extended to${k}_{{\rm{ex}}}+{k}_{{\rm{em}}}$ . (b) Spot-scanning illumination microscopy where fluorescence is collected by an array detector, and pixels offset by a distance from the excitation spot detect a shifted but higher-resolution, low-signal confocal image. The reconstruction algorithm corrects the shift and restores the signal by reassigning the detected fluorescence toward the illumination axis, with the final resolution${PS F}_{{\rm{sys}}}$ determined by the product of the excitation PSF (${PS F}_{{\rm{ex}}}$ ) and the emission PSF (${PS F}_{{\rm{em}}}$ ). After deconvolution, this process improves resolution similar to that obtained with sinusoidal illumination microscopyFigure 4. (a) Schematic diagram of 3D STED-SIM. (b) The cross-section comparison of lateral PSF (top, left), axial PSF (bottom, left), lateral OTF (top, right), and axial OTF (bottom, right) of the widefield microscopy (red) and 3D STED-SIM (blue). Adapted from Xue et al.[49]
Table 1. Comparison of SIM SR reconstruction algorithm
Principle Effect Code Reference TV-SIM Append TV regularization to reconstruction Suppress reconstruction artifacts Not open-source Chu et al. 2014[14] Hessian-SIM Append Hessian regularization to reconstruction Suppress reconstruction artifacts, avoid over-sharpening boundaries Open-source Huang et al. 2018[15] HiFi-SIM Engineering the effective SIM PSF into an ideal form Suppress reconstruction artifacts, improve axial sectioning Open-source Wen et al. 2021[17] Sparse-SIM Append Sparse and Hessian regularization to reconstruction Increases SIM resolution ~2-fold laterally Open-source Zhao et al. 2021[26] sCMOS Noise-corrected SIM Introduce sCMOS imaging noise model to reconstruction Suppress sCMOS noise-induced reconstruction artifacts Not open source Zhou et al. 2022[16] Two-step RL deconvolution SIM Introduce two-step RL deconvolution to reconstruction eliminate ad hoc tuneable parameters Not open source Perez et al. 2016[18] Noise-controlled SIM Introduce a physically realistic noise model to reconstruction Suppress reconstruction artifacts, eliminate ad hoc tuneable parameters, maintain resolution and contrast Open-source Smith et al. 2021[19] GAN TIRF-SIM Use GAN for transforming TIRF images into TIRF SIM images Reconstruct rapidly Open-source Wang et al. 2019[20] U-Net SIM Use U-net for producing SIM images Train efficiently and reconstruct with fewer low-intensity input images Open-source Jin et al. 2020[21] 3D RCAN Use 3D RCAN for increasing SIM resolution Increases SIM resolution ~1.9-fold laterally and ~3.6-fold axially Open-source Chen et al. 2021[23] DFCAN/DFGAN Use DFCAN/DFGAN for producing SIM images Reconstruct with low SNR input images Open-source Qiao et al. 2021[24] Table 2. Summary of SIM performance evaluation algorithms
Function Code Reference FRC/FSC Determine SIM resolution by cross-correlation Open-source Nieuwenhuizen et al. 2013[28] Decorrelation analysis Determine SIM resolution by partial phase correlation Open-source Descloux et al. 2019[31] NanoJ-SQUIRREL Evaluate SIM artifacts with the resolution scaling function Open-source Culley et al. 2018[32] SIMcheck Evaluate SIM stripe modulation contrast by computing the standard deviation Open-source Ball et al. 2015[33] -
[1] BETZIG E, TRAUTMAN J K, HARRIS T D, et al. Breaking the diffraction barrier: optical microscopy on a nanometric scale[J]. Science, 1991, 251(5000): 1468-1470. doi: 10.1126/science.251.5000.1468 [2] AXELROD D. Total internal reflection fluorescence microscopy in cell biology[J]. Traffic, 2001, 2(11): 764-774. doi: 10.1034/j.1600-0854.2001.21104.x [3] HELL S W. Toward fluorescence nanoscopy[J]. Nature Biotechnology, 2003, 21(11): 1347-1355. doi: 10.1038/nbt895 [4] EGGELING C, RINGEMANN C, MEDDA R, et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell[J]. Nature, 2009, 457(7233): 1159-1162. doi: 10.1038/nature07596 [5] BETZIG E, PATTERSON G H, SOUGRAT R, et al. Imaging intracellular fluorescent proteins at nanometer resolution[J]. Science, 2006, 313(5793): 1642-1645. doi: 10.1126/science.1127344 [6] RUST M J, BATES M, ZHUANG X W. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM)[J]. Nature Methods, 2006, 3(10): 793-796. doi: 10.1038/nmeth929 [7] THOMPSON R E, LARSON D R, WEBB W W. Precise nanometer localization analysis for individual fluorescent probes[J]. Biophysical Journal, 2002, 82(5): 2775-2783. doi: 10.1016/S0006-3495(02)75618-X [8] GUSTAFSSON M G L. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy[J]. Journal of Microscopy, 2000, 198(2): 82-87. doi: 10.1046/j.1365-2818.2000.00710.x [9] WU Y, SHROFF H. Faster, sharper, and deeper: structured illumination microscopy for biological imaging[J]. Nature Methods, 2018, 15(12): 1011-1019. doi: 10.1038/s41592-018-0211-z [10] GUSTAFSSON M G L, SHAO L, CARLTON P M, et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination[J]. Biophysical Journal, 2008, 94(12): 4957-4970. doi: 10.1529/biophysj.107.120345 [11] SHROFF S A, FIENUP J R, WILLIAMS D R. Phase-shift estimation in sinusoidally illuminated images for lateral superresolution[J]. Journal of the Optical Society of America A, 2009, 26(2): 413-424. doi: 10.1364/JOSAA.26.000413 [12] WICKER K, MANDULA O, BEST G, et al. Phase optimisation for structured illumination microscopy[J]. Optics Express, 2013, 21(2): 2032-2049. doi: 10.1364/OE.21.002032 [13] ZHOU X, LEI M, DAN D, et al. Image recombination transform algorithm for superresolution structured illumination microscopy[J]. Journal of Biomedical Optics, 2016, 21(9): 096009. doi: 10.1117/1.JBO.21.9.096009 [14] CHU K Q, MCMILLAN P J, SMITH Z J, et al. Image reconstruction for structured-illumination microscopy with low signal level[J]. Optics Express, 2014, 22(7): 8687-8702. doi: 10.1364/OE.22.008687 [15] HUANG X SH, FAN J CH, LI L J, et al. Fast, long-term, super-resolution imaging with Hessian structured illumination microscopy[J]. Nature Biotechnology, 2018, 36(5): 451-459. doi: 10.1038/nbt.4115 [16] ZHOU B, HUANG X SH, FAN J CH, et al. sCMOS noise-corrected superresolution reconstruction algorithm for structured illumination microscopy[J]. Photonics, 2022, 9(3): 172. doi: 10.3390/photonics9030172 [17] WEN G, LI S M, WANG L B, et al. High-fidelity structured illumination microscopy by point-spread-function engineering[J]. Light:Science &Applications, 2021, 10(1): 70. [18] PEREZ V, CHANG B J, STELZER E H K. Optimal 2D-SIM reconstruction by two filtering steps with Richardson-Lucy deconvolution[J]. Scientific Reports, 2016, 6: 37149. doi: 10.1038/srep37149 [19] SMITH C S, SLOTMAN J A, SCHERMELLEH L, et al. Structured illumination microscopy with noise-controlled image reconstructions[J]. Nature Methods, 2021, 18(7): 821-828. doi: 10.1038/s41592-021-01167-7 [20] WANG H D, RIVENSON Y, JIN Y Y, et al. Deep learning enables cross-modality super-resolution in fluorescence microscopy[J]. Nature Methods, 2019, 16(1): 103-110. doi: 10.1038/s41592-018-0239-0 [21] JIN L H, LIU B, ZHAO F Q, et al. Deep learning enables structured illumination microscopy with low light levels and enhanced speed[J]. Nature Communications, 2020, 11(1): 1934. doi: 10.1038/s41467-020-15784-x [22] ZHANG Y L, LI K P, LI K, et al. . Image super-resolution using very deep residual channel attention networks[C]. Proceedings of the 15th European Conference on Computer Vision, Springer, 2018: 294-310. [23] CHEN J J, SASAKI H, LAI H, et al. Three-dimensional residual channel attention networks denoise and sharpen fluorescence microscopy image volumes[J]. Nature Methods, 2021, 18(6): 678-687. doi: 10.1038/s41592-021-01155-x [24] QIAO CH, LI D, GUO Y T, LIU CH, et al. Evaluation and development of deep neural networks for image super-resolution in optical microscopy[J]. Nature Methods, 2021, 18(2): 194-202. doi: 10.1038/s41592-020-01048-5 [25] SCHULZ O, PIEPER C, CLEVER M, et al. Resolution doubling in fluorescence microscopy with confocal spinning-disk image scanning microscopy[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(52): 21000-21005. doi: 10.1073/pnas.1315858110 [26] ZHAO W S, ZHAO SH Q, LI L J, et al. Sparse deconvolution improves the resolution of live-cell super-resolution fluorescence microscopy[J]. Nature Biotechnology, 2022, 40(4): 606-617. doi: 10.1038/s41587-021-01092-2 [27] NIEUWENHUIZEN R P J, LIDKE K A, BATES M, et al. Measuring image resolution in optical nanoscopy[J]. Nature Methods, 2013, 10(6): 557-562. doi: 10.1038/nmeth.2448 [28] BANTERLE N, BUI K H, LEMKE E A, et al. Fourier ring correlation as a resolution criterion for super-resolution microscopy[J]. Journal of Structural Biology, 2013, 183(3): 363-367. doi: 10.1016/j.jsb.2013.05.004 [29] ROSENTHAL P B, HENDERSON R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy[J]. Journal of Molecular Biology, 2003, 333(4): 721-745. doi: 10.1016/j.jmb.2003.07.013 [30] SAXTON W O, BAUMEISTER W. The correlation averaging of a regularly arranged bacterial cell envelope protein[J]. Journal of Microscopy, 1982, 127(2): 127-138. doi: 10.1111/j.1365-2818.1982.tb00405.x [31] DESCLOUX A, GRUßMAYER K S, RADENOVIC A. Parameter-free image resolution estimation based on decorrelation analysis[J]. Nature Methods, 2019, 16(9): 918-924. doi: 10.1038/s41592-019-0515-7 [32] CULLEY S, ALBRECHT D, JACOBS C, et al. Quantitative mapping and minimization of super-resolution optical imaging artifacts[J]. Nature Methods, 2018, 15(4): 263-266. doi: 10.1038/nmeth.4605 [33] BALL G, DEMMERLE J, KAUFMANN R, et al. . SIMcheck: a toolbox for successful super-resolution structured illumination microscopy[J]. Scientific Reports, 2015, 5: 15915. [34] FIOLKA R, BECK M, STEMMER A. Structured illumination in total internal reflection fluorescence microscopy using a spatial light modulator[J]. Optics Letters, 2008, 33(14): 1629-1631. doi: 10.1364/OL.33.001629 [35] KNER P, CHHUN B B, GRIFFIS E R, et al. Super-resolution video microscopy of live cells by structured illumination[J]. Nature Methods, 2009, 6(5): 339-342. doi: 10.1038/nmeth.1324 [36] LI D, SHAO L, CHEN B CH, et al. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics[J]. Science, 2015, 349(6251): aab3500. doi: 10.1126/science.aab3500 [37] NIXON-ABELL J, OBARA C J, WEIGEL A V, et al. Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER[J]. Science, 2016, 354(6311): aaf3928. doi: 10.1126/science.aaf3928 [38] GUO Y T, LI D, ZHANG S W, et al. Visualizing intracellular organelle and cytoskeletal interactions at nanoscale resolution on millisecond timescales[J]. Cell, 2018, 175(5): 1430-1442.e17. doi: 10.1016/j.cell.2018.09.057 [39] YORK A G, PAREKH S H, NOGARE D D, et al. Resolution doubling in live, multicellular organisms via multifocal structured illumination microscopy[J]. Nature Methods, 2012, 9(7): 749-754. doi: 10.1038/nmeth.2025 [40] YORK A G, CHANDRIS P, NOGARE D D, et al. Instant super-resolution imaging in live cells and embryos via analog image processing[J]. Nature Methods, 2013, 10(11): 1122-1126. doi: 10.1038/nmeth.2687 [41] MO Y Q, FENG F, MAO H, et al. Structured illumination microscopy artefacts caused by illumination scattering[J]. Philosophical Transactions of the Royal Society A:Mathematical,Physical and Engineering Sciences, 2021, 379(2199): 20200153. doi: 10.1098/rsta.2020.0153 [42] INGARAMO M, YORK A G, WAWRZUSIN P, et al. Two-photon excitation improves multifocal structured illumination microscopy in thick scattering tissue[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(14): 5254-5259. doi: 10.1073/pnas.1314447111 [43] WINTER P W, YORK A G, NOGARE D D, et al. Two-photon instant structured illumination microscopy improves the depth penetration of super-resolution imaging in thick scattering samples[J]. Optica, 2014, 1(3): 181-191. doi: 10.1364/OPTICA.1.000181 [44] GREGOR I, SPIECKER M, PETROVSKY R, et al. Rapid nonlinear image scanning microscopy[J]. Nature Methods, 2017, 14(11): 1087-1089. doi: 10.1038/nmeth.4467 [45] HEINTZMANN R, JOVIN T M, CREMER C. Saturated patterned excitation microscopy-a concept for optical resolution improvement[J]. Journal of the Optical Society of America A, 2002, 19(8): 1599-1609. doi: 10.1364/JOSAA.19.001599 [46] GUSTAFSSON M G L. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(37): 13081-13086. doi: 10.1073/pnas.0406877102 [47] ZHANG H, ZHAO M, PENG L L. Nonlinear structured illumination microscopy by surface plasmon enhanced stimulated emission depletion[J]. Optics Express, 2011, 19(24): 24783-24794. doi: 10.1364/OE.19.024783 [48] DAKE F, NAKAYAMA S, TAKI Y. Optical resolution enhancement and background reduction by stimulated emission depletion structured illumination microscopy with structured excitation[C]. Novel Techniques in Microscopy 2015, OSA, 2015: NM2C. 4. [49] XUE Y, SO P T C. Three-dimensional super-resolution high-throughput imaging by structured illumination STED microscopy[J]. Optics Express, 2018, 26(16): 20920-20928. doi: 10.1364/OE.26.020920 [50] REGO E H, SHAO L, MACKLIN J J, et al. Nonlinear structured-illumination microscopy with a photoswitchable protein reveals cellular structures at 50-nm resolution[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(3): E135-E143. -














 下载:
下载: