2022 Vol. 15, No. 6
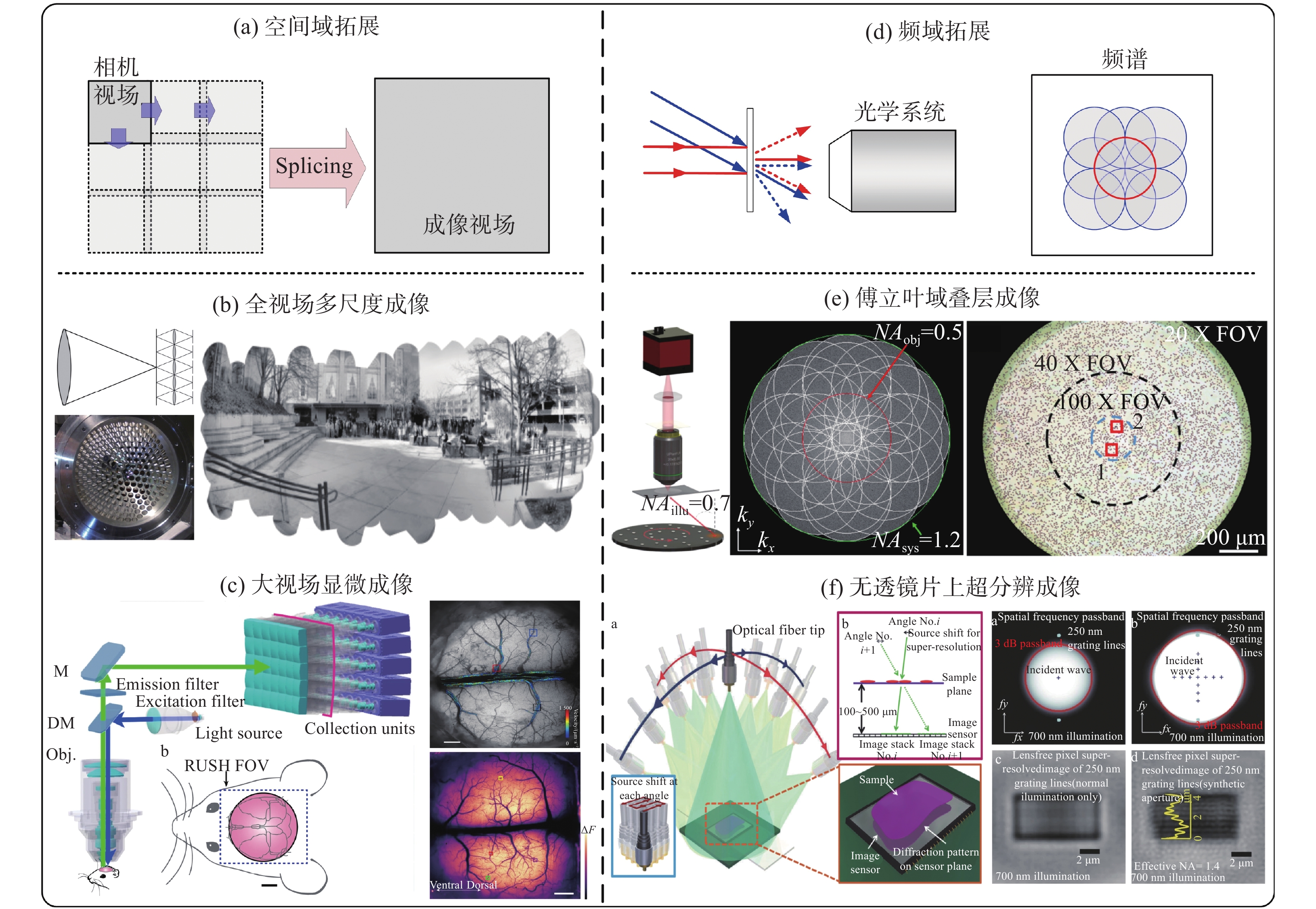
Conventional optical imaging is essentially a process of recording and reproducing the intensity signal of a scene in the spatial dimension with direct uniform sampling. In this process, the resolution and information content of imaging are inevitably constrained by several physical limitations such as optical diffraction limit, detector sampling, and spatial bandwidth product of the imaging system. How to break these physical limitations and obtain higher resolution and broader image field of view has been an eternal topic in this field. In this paper, we introduce the basic theories and technologies associated with the resolution, super-resolution, and spatial bandwidth product expansion, as well as some examples in the field of computational optical imaging. By placing these specific cases into the higher dimensional framework of "computational optical imaging", this paper reveals that most of them can be understood as a "spatial bandwidth regulation" scheme, i.e., a process of exploiting the available degrees of freedom of the imaging system to optimally encode, decode, and transmit information within the constraints of the limited spatial bandwidth of the imaging system, or figuratively speaking - "dancing with shackles". This is essentially a legal trade-off and choice between "gain" and "loss" under physical constraints. The conclusions of this paper are expected to provide valuable insights into the design and exploration of new imaging mechanisms and methods for various complex practical imaging applications.
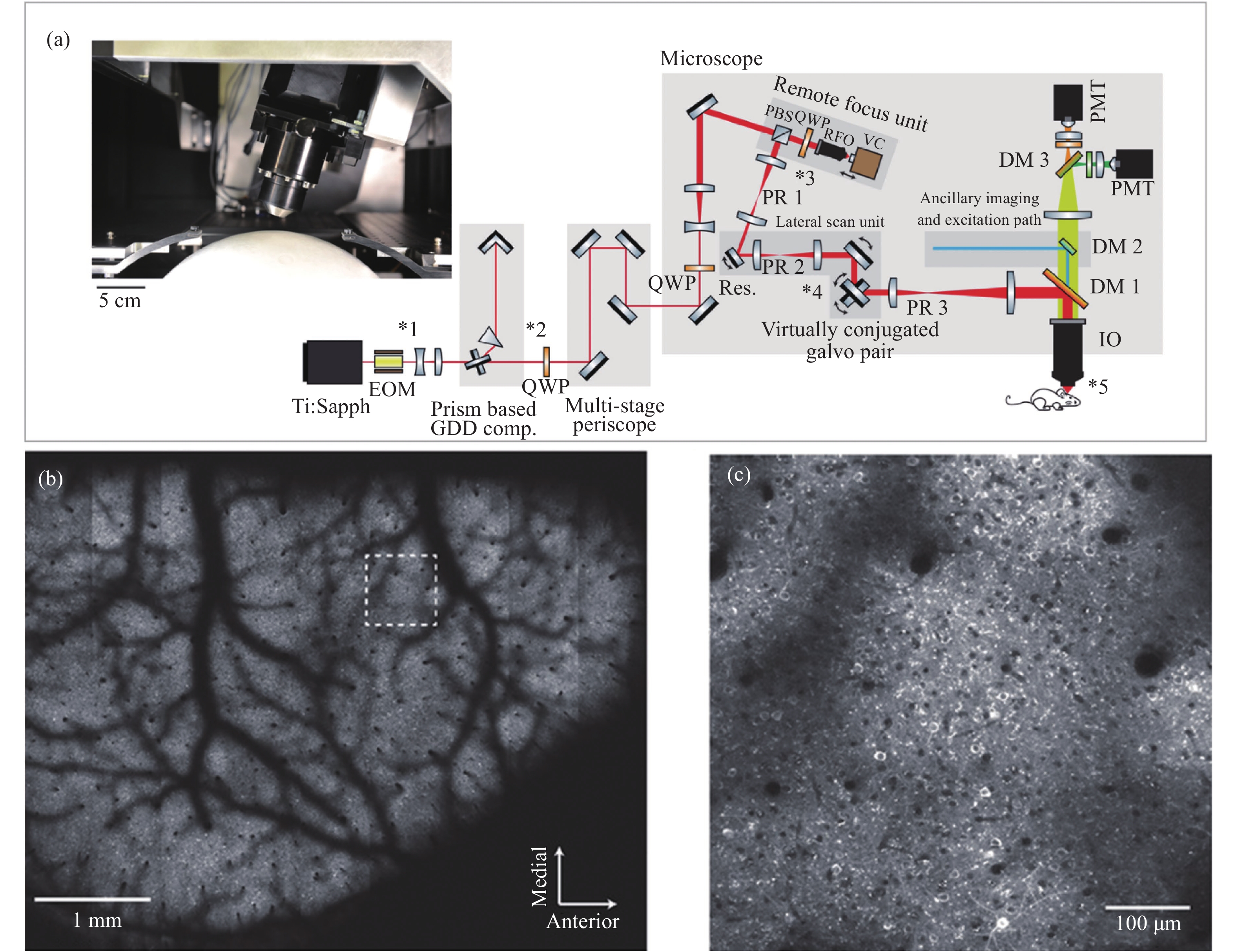
Two-photon microscopy’s ability to maintain good spatial resolution in thick biological tissues has led to its application in in-vivo brain imaging studies soon after its conception. As neural networks have cross-scale multidimensional spatio-temporal properties, two-photon microscopy has developed rapidly and significantly in recent years to meet the demand for in-vivo cross-scale imaging of the brain. This paper firstly introduces the working principle of two-photon microscopy, then reviews the progress of two-photon microscopy from five perspectives: imaging field of view, imaging flux, imaging depth, resolution, miniaturization, and analyzes the difficulties and future challenges of cross-scale two-photon in-vivo microscopic imaging technology.
Optical imaging has become the dominant method for characterizing information in biological systems. The rapid, non-destructive and comprehensive characterization of biological samples in recent years has placed high demands on the resolvable volume of imaging systems. Digital holography records an entire complex wavefront including both the amplitude and phase of the light field by interference imaging. Due to fast, non-destructive, and 3D imaging abilities, digital holography has been used in numerous applications such as digital pathology, label-free observation and real-time monitoring of in vitro cells. First, this paper introduces the main ways to achieve high-throughput imaging, and analyzes the advantages of digital holography and the evolution of spatial bandwidth. Secondly, a theoretical framework for high-throughput multi-channel multiplexing digital holography based on the Hilbert transform is presented. Then, an extended field of view digital holographic microscope is introduced based on this theoretical framework. Experimental results indicate that the system achieves 8 times the space-bandwidth product higher than that of conventional off-axis holographic microscopes without sacrificing spatial and temporal resolution. This high-throughput digital holographic multiplexing technology can make full use of the redundant spatial bandwidth of single intensity image, which verifies the feasibility of high-throughput multi-channel multiplexing digital holography.
With the characteristics of real-time, high-resolution and non-invasive, optical microscopy can scale from cells, tissues to whole living organisms, which has greatly expanded our understanding to the nature of life. However, due to the limited Space-Bandwidth Product (SBP), it is hard for a conventional optical microscope to achieve a large field of view with a high resolution. This makes it very difficult for microscopic imaging in large field of view biological imaging applications, such as imaging of neural circuits between the synapse of the brain neural networks. Recently, large field-of-view imaging technology has received increasing attention and experienced rapid development. The SBP has been improved ten times or even a hundred times as compared to a traditional optical microscope and the field-of-view has been expanded without sacrificing resolution, which, in turn, has resolved some major problems in biomedical research. This review introduces the progress, characteristics and corresponding biological applications of several typical trans-scale optical imaging techniques in recent years, and gives an outlook on their future development.
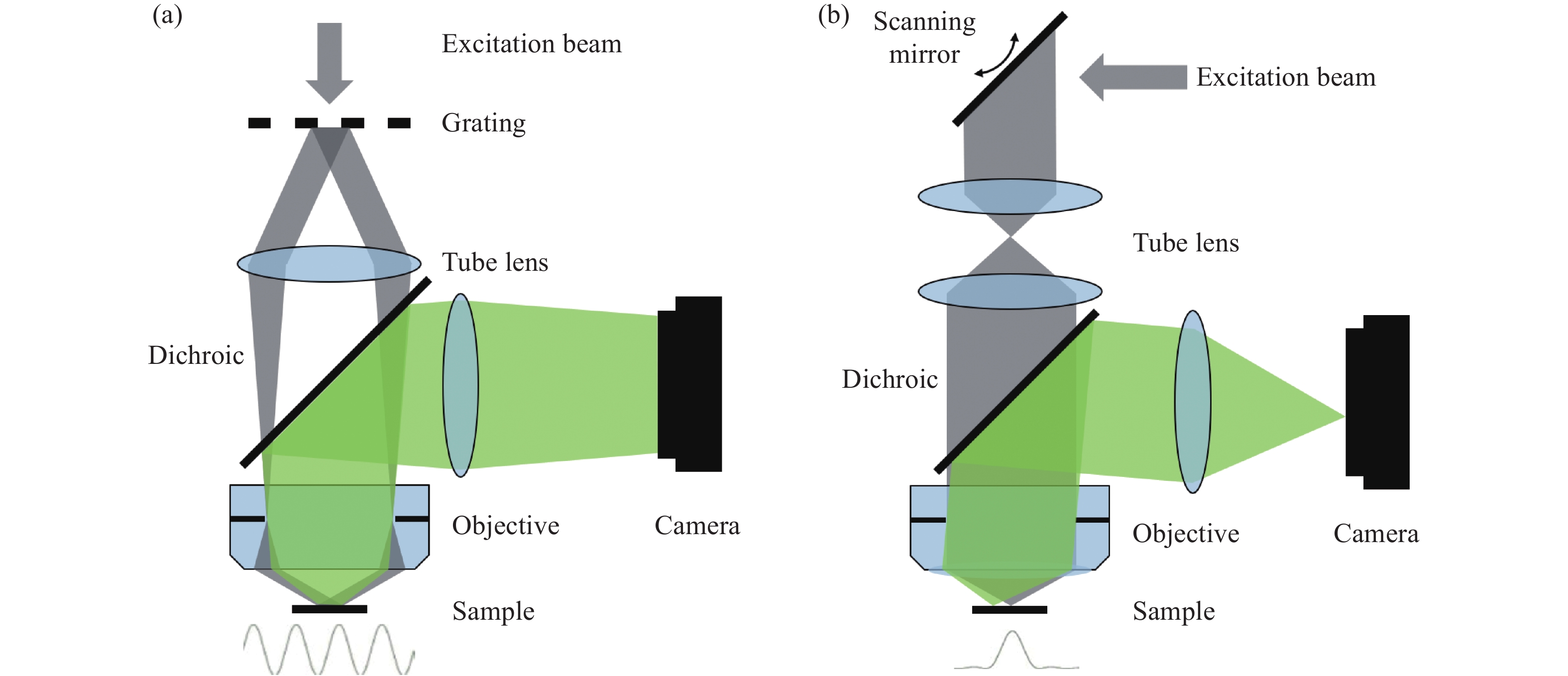
As an early component of modern Super-Resolution (SR) imaging technology, Structured Illumination Microscopy (SIM) has been developed for nearly twenty years. With up to ~60 nm wavelengths and 564 Hz frame rates, it has recently achieved an optimal combination of spatiotemporal resolution in live cells. Despite these advantages, SIM also suffers disadvantages, some of which originated from the intrinsic reconstruction process. Here we review recent technical advances in SIM, including SR reconstruction, performance evaluation, and its integration with other technologies to provide a practical guide for biologists.
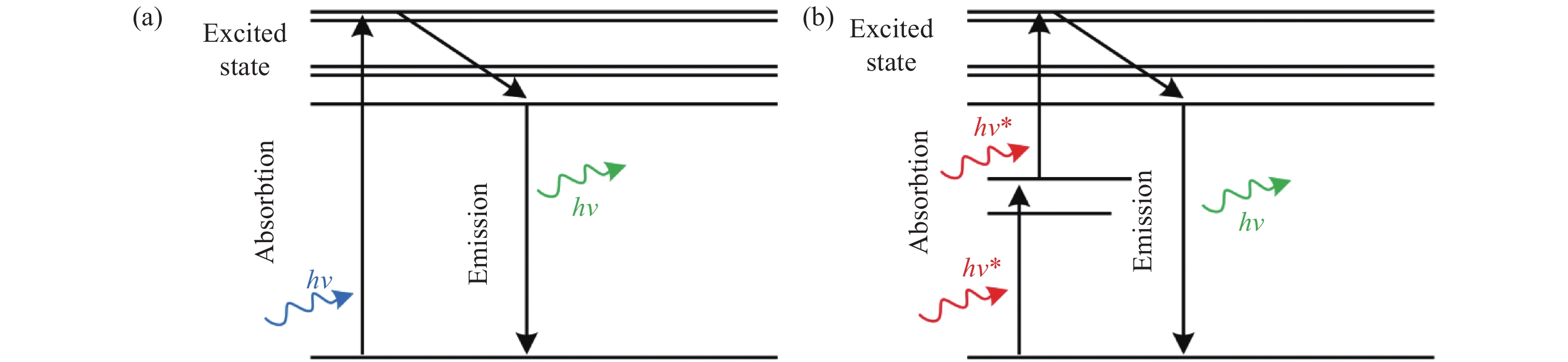
Lipid droplets are a kind of spherical organelle in eukaryotic cells and are relevant to many cellular physiological processes. Fluorescence imaging techniques are one of the most powerful tools to visualize and study lipid droplets. However, conventional wide-field microscopy and confocal microscopy can only provide a resolution of about 250 nm due to the limitation of optical diffraction. This resolution is quite insufficient for visualizing the small lipid droplets, especially the nascent ones (size of about 30~60 nm). Emerging super-resolution microscopes that can break the diffraction limit (such as stimulated emission depletion microscopy, structured illumination microscopy and photoactivated localization microscopy) have gradually attracted much interest in recent years. To obtain high-resolution fluorescence images of lipid droplets, the advanced fluorescent probes which meet the special requirements of the corresponding super-resolution microscopes are highly essential. This review paper will briefly introduce the working principles of various super-resolution microscopes, discuss the special requirements on the photophysical properties of fluorescent probes, and systematically summarize the research progress of super-resolution imaging of lipid droplets by employing these fluorescent probes. Meanwhile, this review will compare the advantages and shortcomings of different super-resolution techniques for lipid droplets imaging, and prospect their future possible trends.
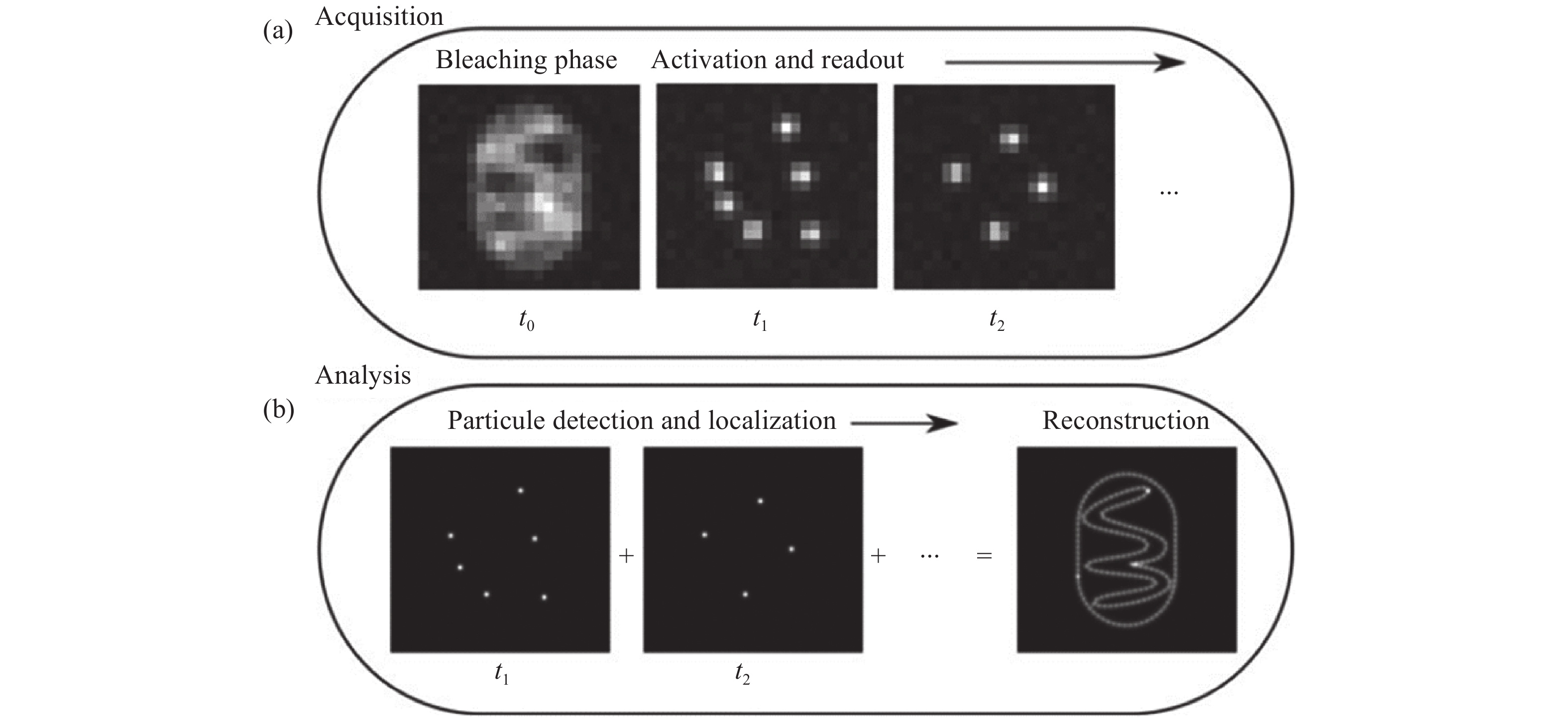
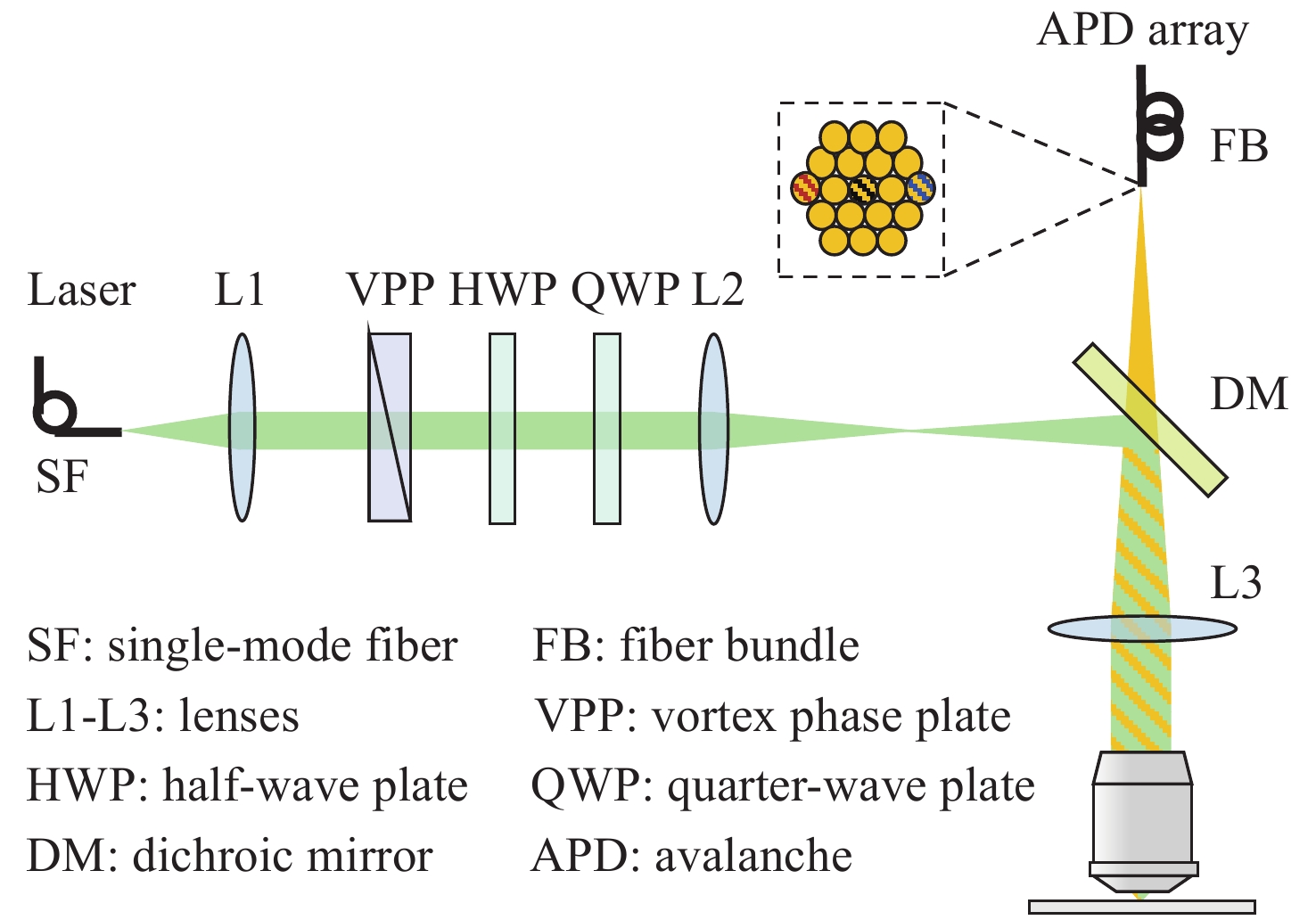
Single-molecule imaging is widely used for the reconstruction of three-dimensional subcellular structures. The point spread function is an important window to analyze the information of a single molecule. Besides 3D coordinates, it also contains abundant additional information. In this paper, we reviewed the recent progress of multi-dimensional single-molecule imaging, including spatial location, fluorescence wavelength, dipole orientation, interference phase, etc. We also briefly introduced the latest methods for molecule localization and proposed the further directions for its research.
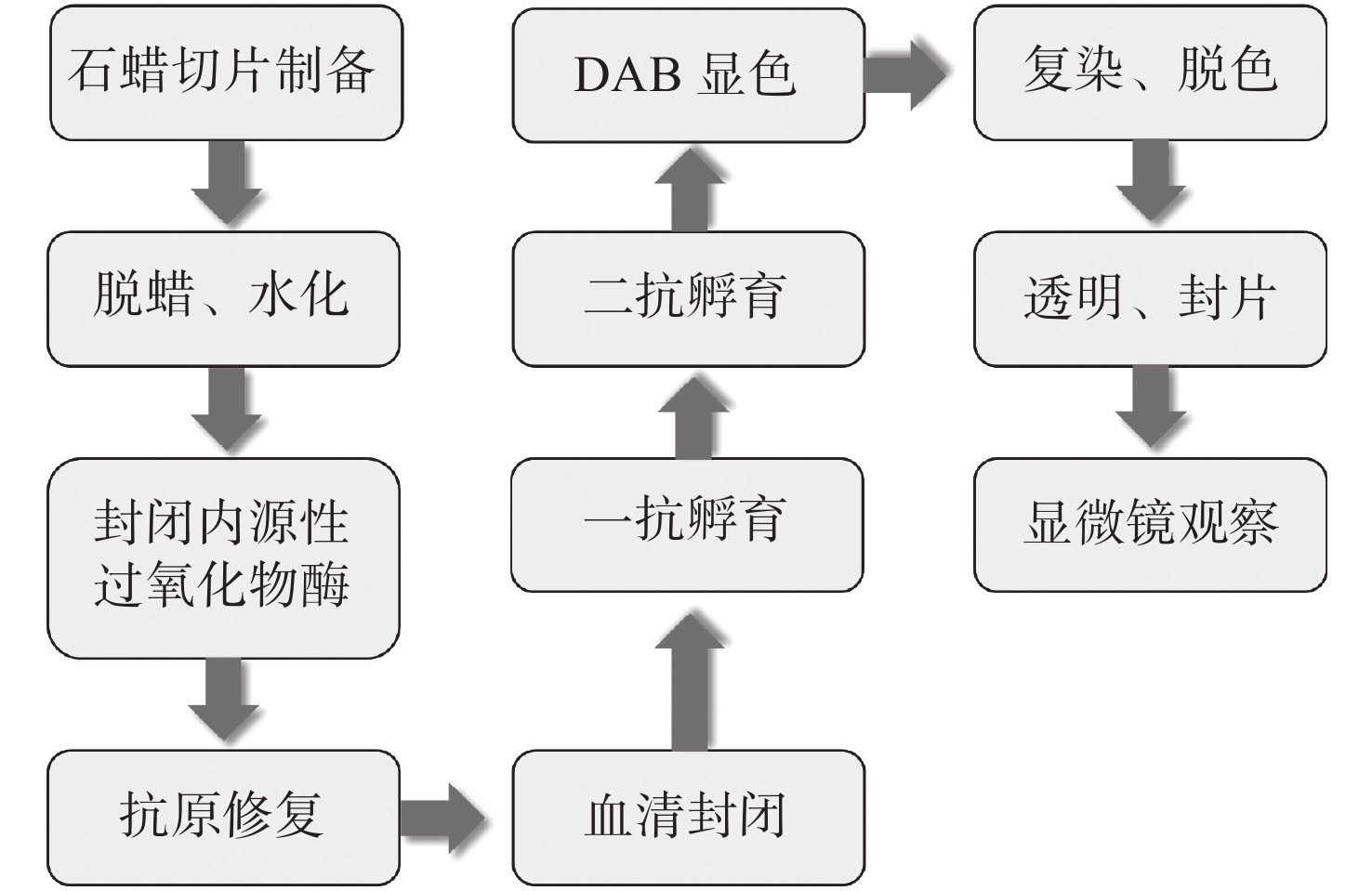
Digital pathology has brought new opportunities for remote pathological consultation and joint consultation owing to its convenient storage, management, browsing and transmission. However, because of the limited field of view of a microscope, panoramic imaging cannot be achieved while ensuring a high resolution. The proposal of panoramic digital pathology makes up for this defect and achieves panoramic imaging while ensuring high resolution. However, a single slice can only detect a single target, and disease diagnosis needs to observe the expression of multi-target at the same time. In recent years, multi-target panoramic digital pathology technology has developed rapidly. It has attracted much attention because of its great application potential in drug research and development, clinical research and basic research. Owing to its large field of view, wide range of colors and high flux, the system can detect the expression of various biomarkers on a whole tissue section in situ in a short time to identify the phenotype, abundance, state, and relationship of each cell. Firstly, this paper reviews the development process of digital pathology, panoramic digital pathology and multi-target panoramic digital pathology, as well as the update and iteration of technology in the development process, and illustrates the importance of developing multi-target panoramic digital pathology. Then, the multi-target panoramic digital pathology is described in detail from three perspectives: biological sample preparation, multi-color imaging system and image processing. Next, the applications of multi-target panoramic digital pathology in biomedical fields, such as tumor microenvironments and tumor molecular typing are described. Finally, the advantages, challenges and future development of multi-target panoramic digital pathology are summarized.
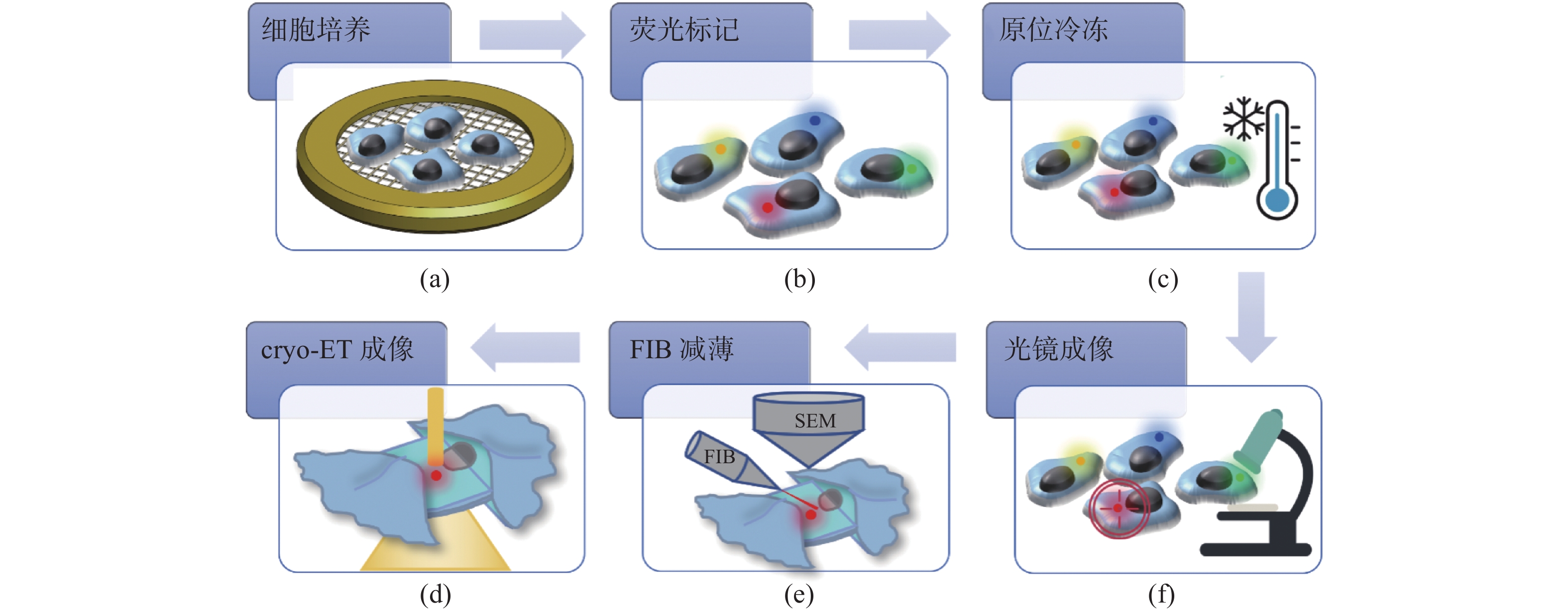
Cryo-electron tomography (cryo-ET) has become a cutting-edge technology in life sciences for the investigation of protein complexes directly in their natural state. In cryo-ET, the sample’s thickness must be less than 300 nm and the target molecule must be within the lamella, which is prepared by cryo-Focus Iron Beam (FIB) milling. In order to precisely navigate molecules and to improve the efficiency of sample preparation, cryo-Correlative Light and Electron Microscopy (cryo-CLEM) has been introduced to perform in-situ imaging on the frozen samples. The cryo-CLEM combines the localization advantages of fluorescence imaging with the resolution advantages of electron microscopy. By registering images of light and electrons, frozen samples can be thinned by FIB milling, so the efficiency of cryo-ET sample preparation can be improved. In this paper, we review the latest progress and applications of cryo-CLEM technologies, with a particular focus on super-resolution cryo-CLEM imaging and integrated cryo-CLEM. The advantages and limitations of various methodologies, as well as their application scope, are discussed. A discussion on cryo-CLEM's limitations and potential directions for its future development are also presented.
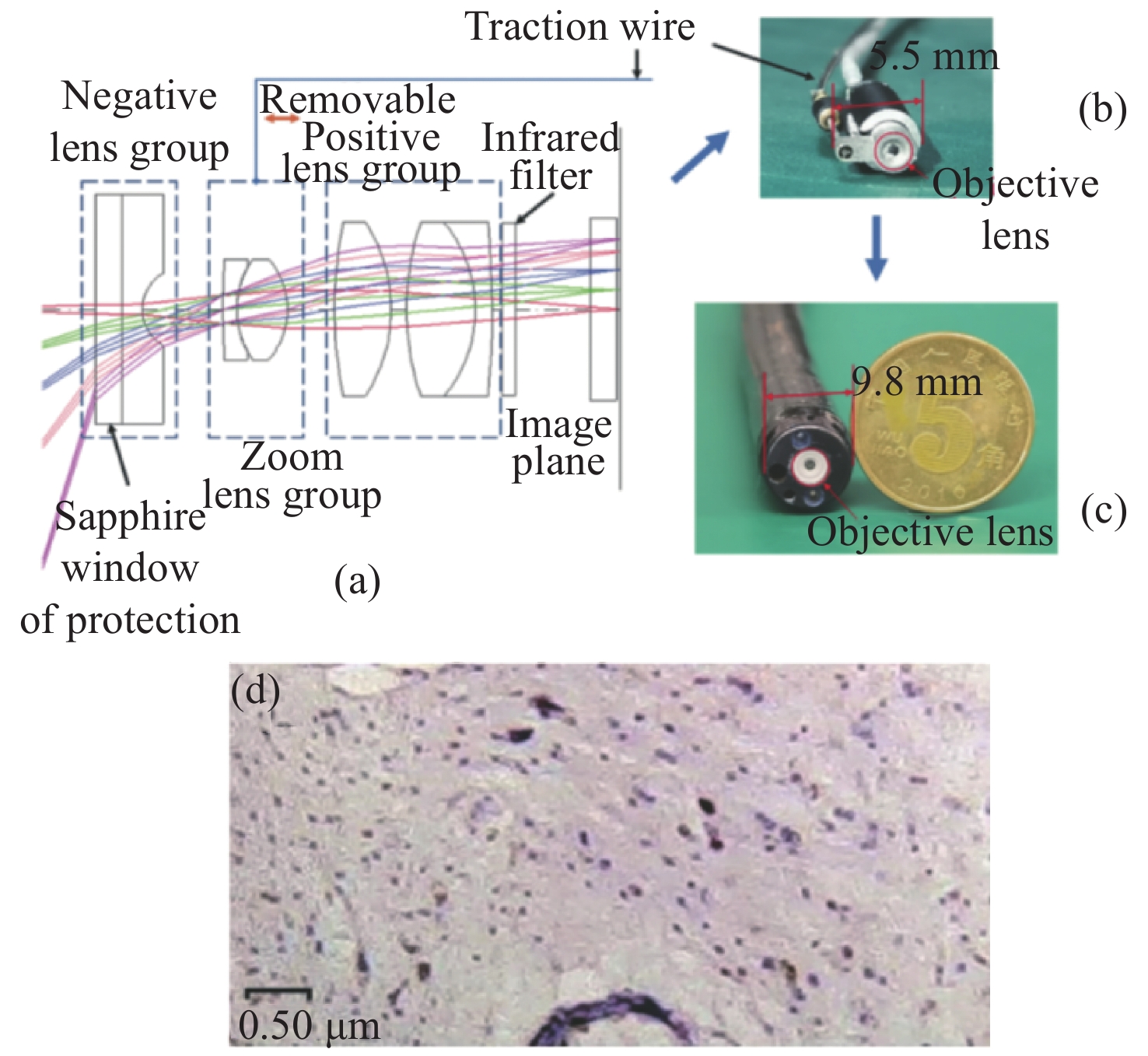
Due to the advantages of high resolution, multi-scale, multi-dimension, low radiation and easy to integrate, optical imaging technology plays an important role in biomedical field. In the field of endoscopy, how to obtain, process and visualize the endoscopic image information is the core of the problem what optical imaging technology need to solve. The obtaining of trans-scale endoscopic image of patients in the medical clinical is more advantageous to the surgeon for the diagnosis of patients and can improve in accuracy of the operation. The review starts with the application of trans-scale optical imaging technology in the field of endoscopy, focusing on the different optical systems to obtain trans-scale images in clinical endoscopy, including trans-scale optical zoom system, multi-channel imaging system, fiber-scanning imaging system, and expounds its progress and future trends.
The fields of modern biology and biomedicine urgently need wide-field-of-view (FOV), high-resolution microscopic technology and instruments for trans-scale observation of biological samples to meet the requirement of major scientific for research. Limited by the spatial bandwidth product, traditional commercial microscopes cannot meet this demand. Besides, the existing high spatial bandwidth product microscopy systems have problems such as bulky volume and high implementation costs. In this paper, based on the HiLo optical sectioning technology and the self-designed wide-field-of-view and high-resolution objective, a wide-field-of-view and high-resolution HiLo optical sectioning microscopy system was developed. The FOV and imaging resolution of this system were tested. Brightfield imaging experiments were carried out on mouse brain slices by this system and the results were compared with that of OLYMPUS commercial microscope. At the same time, wide-field fluorescence imaging comparison experiments were carried out on wheat seed fluorescent slices. The experiment results show that the FOV of this system reaches 4.8 mm×3.6 mm (the diagonal FOV is 6.0 mm), the lateral resolution reaches 0.74 μm, and the axial resolution reaches 4.16 μm. The comparative experiment proved that this system has the advantages of wide FOV, high resolution and the ability of fast optical sectioning imaging simultaneously. This system can carry out rapid 3D imaging of large-volume biological samples, which will provide strong technical support for researches such as embryonic development, brain imaging, and digital pathology diagnosis.
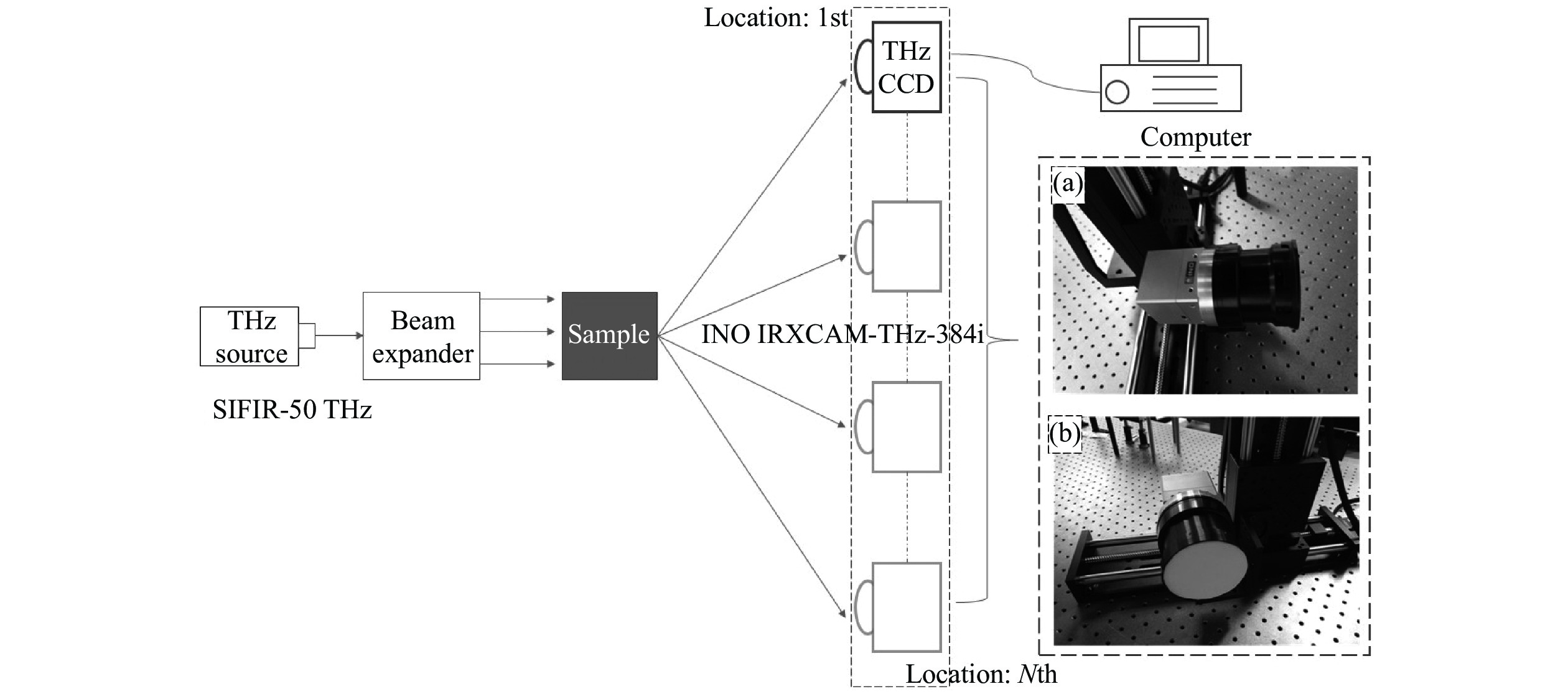
Terahertz (THz) technology becomes increasingly important nowadays, especially in testing and security applications. Extending the field of view and increasing the imaging quality are both vital challenges for THz imaging. To address these problems, we build a THz light field imaging system based on a single-camera scanning configuration, which utilizes the 4D information of the spatial and angular distribution of THz waves. Based on the 4D plenoptic function and the parameterization method with two parallel planes, the intensity consistency of THz propagation is used for refocusing calculation, then a series of refocused images can be obtained by integrating original light field images corresponding to different imaging distances and views. Compared with the original THz imaging, the field of view and the imaging quality of the THz light field imaging are effectively improved. In our experiment, the field of view was enlarged by a factor of 1.84 and the resolution increased from 1.3 mm to 0.7 mm. Furthermore, information on some obscured targets could also be retrieved by defocusing the obstructions. This method could improve the imaging quality of THz imaging as well as expand its functions, which inspires a new way for THz nondestructive testing (NDT) and security inspection.
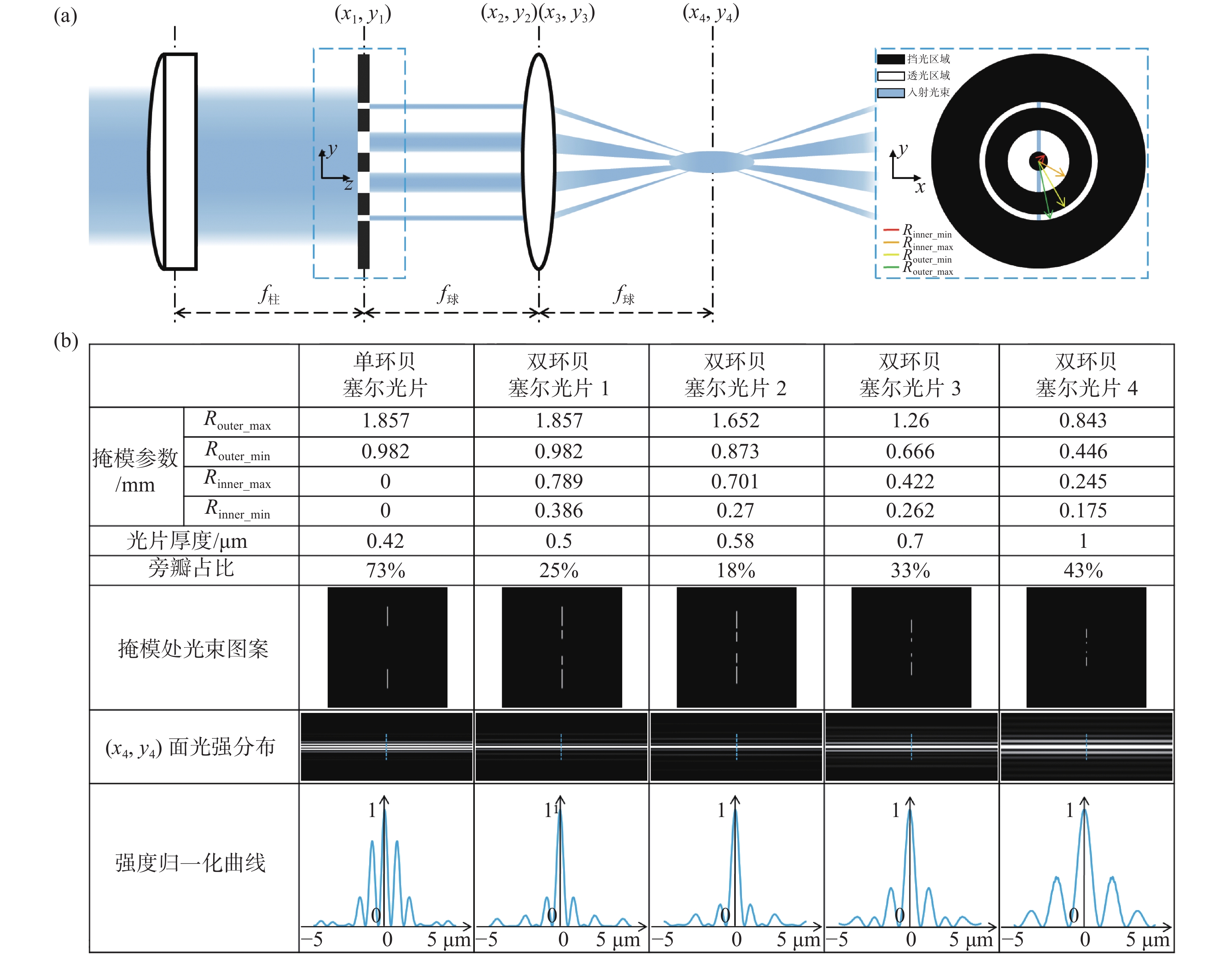
In this paper, we propose a non-diffraction Light Sheet Fluorescence Microscopy (LSFM) technique, which readily enables multi-scale 3D fluorescence imaging of diverse biological samples with size ranging from microns to centimeters. To solve the problem of heavy sidelobes in conventional non-diffraction Bessel LSFM, we invent a double-ring-modulated approach which can generate non-diffraction light sheets with ~0.4 to ~5 µm tunable thickness and the ratio of the sidelobe was reduced to less than 30%. Then we built a multi-scale LSFM system based on this novel approach. The system showed versatile multi-scale imaging abilities, such as dual-color 3D dynamic imaging of single live cell, 3D super-resolution imaging of expansion cells and high-throughput 3D mapping of entire meso-scale organs. Therefore, we demonstrate that this multi-scale imaging modality can substantially improve the efficiency of LSFM for advancing various biomedical studies, such as cell biology, tissue pathology, and neuroscience.
Fluorescence emission difference microscopy is a super-resolution imaging technique with strong universality of fluorescent dyes and low phototoxicity. However, due to the limitation of its principle, traditional fluorescence emission difference microscopy has a high system complexity, low stability and limited imaging speed. In order to improve these defects, we design and build a set of multi-color virtual fluorescence difference microscopy system, and it’s imaging method and parameter are analyzed. On the basis of the existing principle of multi-color virtual fluorescence emission difference microscopy, the influence of the signal-to-noise ratio and background is further considered, and a virtual fluorescence emission difference microscopy imaging model that can be verified experimentally is established. The experiments show that the system and method have the characteristics of simple structure, strong background denoising ability, strong universality of fluorescent dyes, and low phototoxicity. Its imaging resolution is 1.9 times higher than that of confocal system, and its imaging speed is doubled compared to the traditional fluorescence emission difference microscopy system. It has obtained good imaging results at three wavelengths, and has been experimentally verified in biological cell imaging.
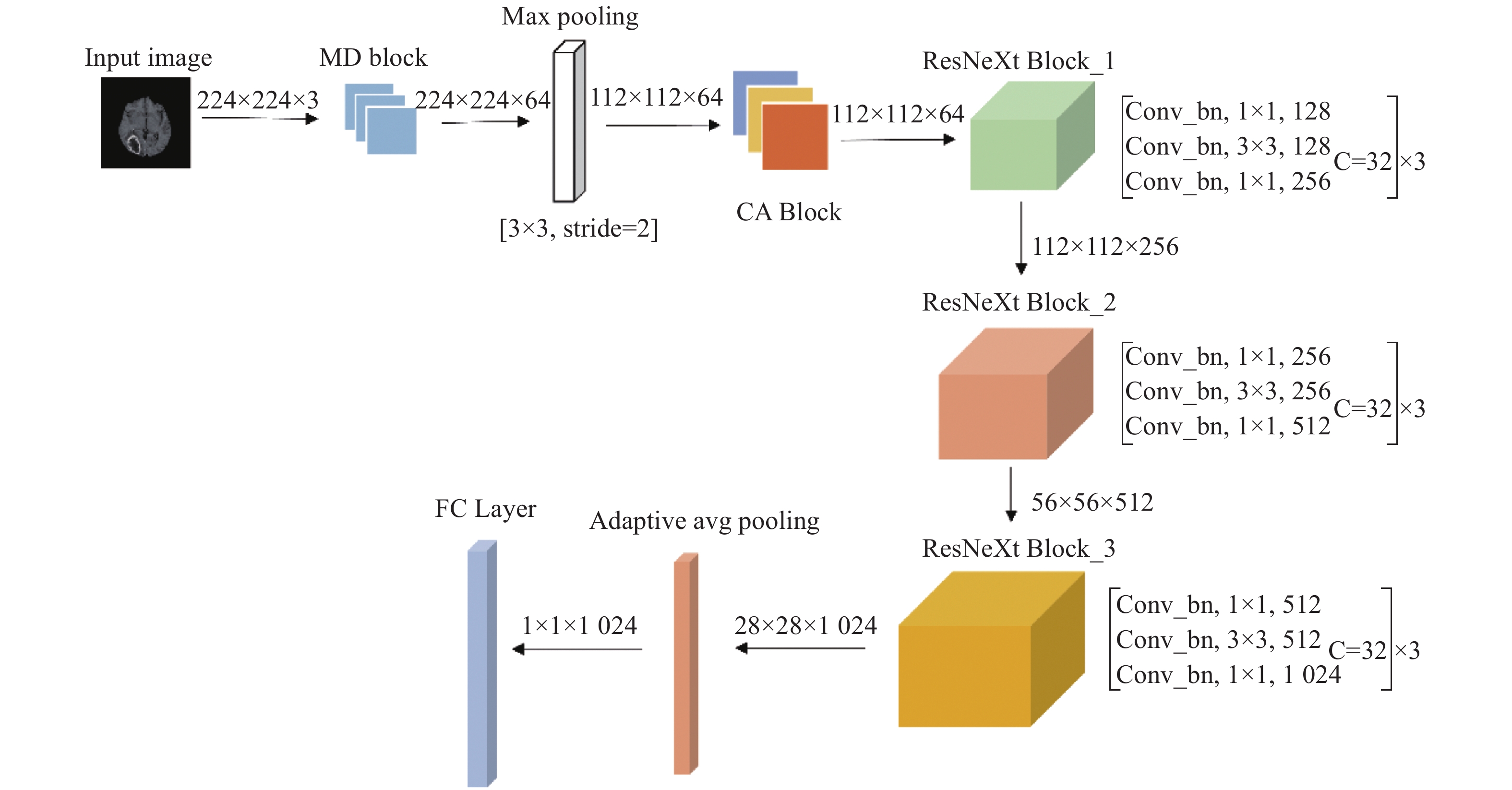
Aiming at the problems of complex and inaccurate classification of benign and malignant brain tumors, a classification model was proposed based on the fusion of multi-scale and channel features. The model used ResNeXt as the backbone network. First, the multi-scale feature extraction module based on dilated convolution was used to replace the first convolution layer, which can make full use of dilation rates to obtain the image information from different receptive fields, and combine the global features with significant subtle ones. Second, the channel attention mechanism module was added in the network to fuse the feature channel information in order to increase the attention to the tumor, and reduce the attention to redundant information. Finally, the combination optimization strategy, the MultiStepLR strategy of the learning rate, the label smoothing strategy and the transfer learning strategy on medical images were adopted to improve the learning and generalization abilities of the model. The experiments were carried out on BraTS2017 Dataset and BraTS2019 Dataset, and the classification accuracy were 98.11% and 98.72%, respectively. Compared with other advanced methods and classical models, the proposed classification model can effectively reduce the complexity of the classification process and improve the detection accuracy of benign and malignant brain tumors.
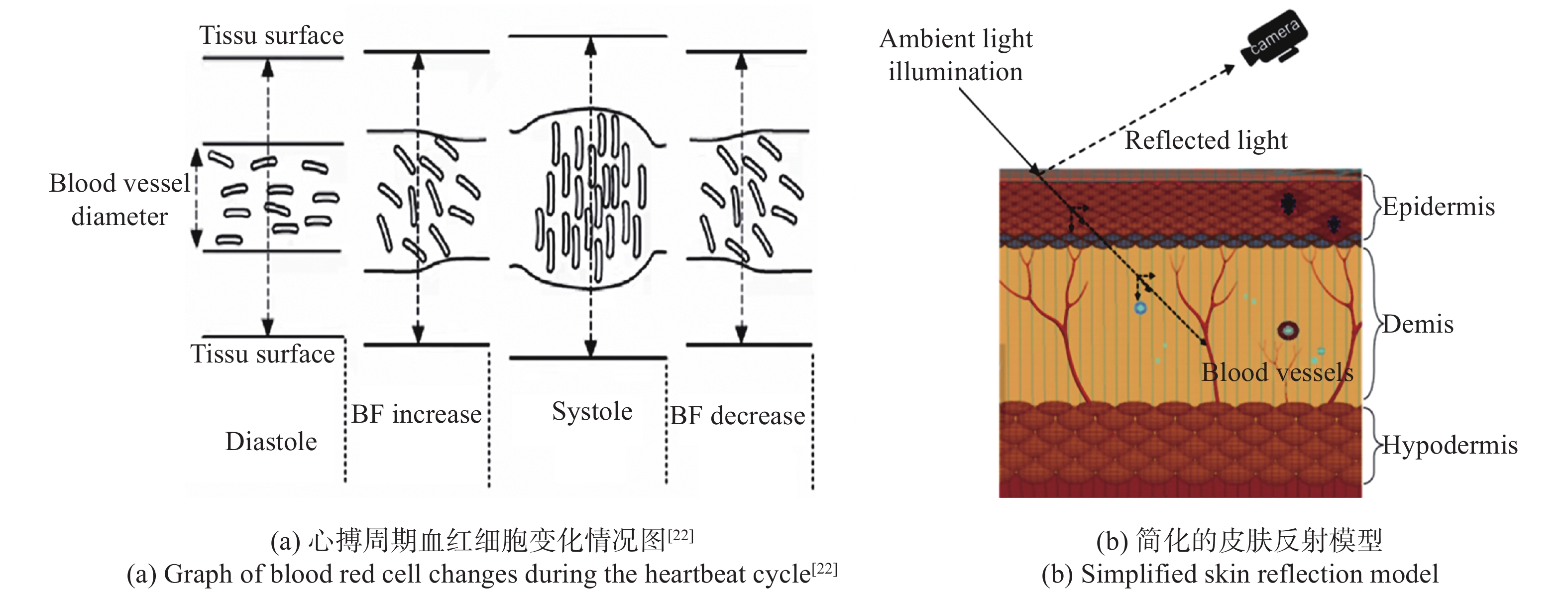
To achieve non-contact daily mental stress detection, this paper proposes a image photoplethysmography to detect mental stress. First, a video of the subject's face is recorded by the cell phone camera. Then, the proposed Dynamic Region of Interest (ROI) extraction method based on Face Mesh is used to obtain the weak skin color changes caused by heart rate fluctuations. Next, the Fast Independent Component Analysis (FastICA) algorithm, wavelet transform and narrowband bandpass filtering are combined to extract the signal and heart rate variability information based on image photoplethysmography. Then, stress-induced experiments are conducted on 30 subjects to screen 14 features for mental stress detection by comparing the differences in heart rate variability parameters between normal and stressful states, and to explore the relationship between short-term mental stress and daily mental stress due to stress induction. Finally, an additional 67 subjects are tested for daily mental stress, and a triple classifier for mental stress detection is built using the machine learning algorithm. The experimental results show that the accuracy of the three classifications of mental stress can reach 95.2%. Given that this method does not require long-term measurements and can accurately detect human mental stress levels using only a smartphone, and that the measurement method is simple, and easy to administer, and does not affect the normal psychological and mental state of the subject, it can be used as a valid tool in psychological research.


 Abstract
Abstract FullText HTML
FullText HTML PDF 19381KB
PDF 19381KB